Origin and evolution of LX4 genotype infectious bronchitis coronavirus in China
- PMID: 28062013
- PMCID: PMC7117135
- DOI: 10.1016/j.vetmic.2016.11.014
Origin and evolution of LX4 genotype infectious bronchitis coronavirus in China
Abstract
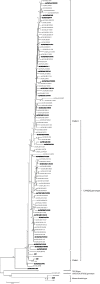
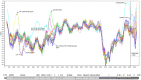
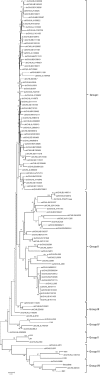
We investigated the genomic characteristics of 110 LX4 genotype strains of infectious bronchitis viruses (IBVs) isolated between 1995 and 2005 in China. The genome of these IBVs varies in size from 27596bp to 27790bp. Most IBV strains have the typical genomic organization of other gamacoronaviruses, however, two strains lacked 3a and 5b genes as a result of a nucleotide change within the start codon in the 3a or 5b genes. Analysis of our 110 viruses revealed that recombination events may be responsible for the emergence of the LX4 genotype with different topologies. Most of these viruses disappeared (before mid-2005) because they were not "fit" to adaptation in chickens. Finally, those of the "fit" viruses (after mid-2005) continued to evolve and have become widespread and predominant in commercial poultry. In addition, few of these viruses experienced recombination with those of the vaccine strains at the 3' end of the genome.
Keywords: Clade I; Clade II; Evolution; Infectious bronchitis virus (IBV); LX4 genotype (QX-like); Topology.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures


References
-
- Boursnell M.E., Brown T.D., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. - PubMed
-
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. - PubMed
-
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

