Mycobacterium tuberculosis Complex Exhibits Lineage-Specific Variations Affecting Protein Ductility and Epitope Recognition
- PMID: 28062754
- PMCID: PMC5521731
- DOI: 10.1093/gbe/evw279
Mycobacterium tuberculosis Complex Exhibits Lineage-Specific Variations Affecting Protein Ductility and Epitope Recognition
Abstract
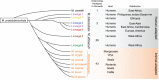
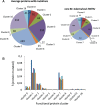
The advent of whole-genome sequencing has provided an unprecedented detail about the evolution and genetic significance of species-specific variations across the whole Mycobacterium tuberculosis Complex. However, little attention has been focused on understanding the functional roles of these variations in the protein coding sequences. In this work, we compare the coding sequences from 74 sequenced mycobacterial species including M. africanum, M. bovis, M. canettii, M. caprae, M. orygis, and M. tuberculosis. Results show that albeit protein variations affect all functional classes, those proteins involved in lipid and intermediary metabolism and respiration have accumulated mutations during evolution. To understand the impact of these mutations on protein functionality, we explored their implications on protein ductility/disorder, a yet unexplored feature of mycobacterial proteomes. In agreement with previous studies, we found that a Gly71Ile substitution in the PhoPR virulence system severely affects the ductility of its nearby region in M. africanum and animal-adapted species. In the same line of evidence, the SmtB transcriptional regulator shows amino acid variations specific to the Beijing lineage, which affects the flexibility of the N-terminal trans-activation domain. Furthermore, despite the fact that MTBC epitopes are evolutionary hyperconserved, we identify strain- and lineage-specific amino acid mutations affecting previously known T-cell epitopes such as EsxH and FbpA (Ag85A). Interestingly, in silico studies reveal that these variations result in differential interaction of epitopes with the main HLA haplogroups.
Keywords: Mycobacterium; coding sequences; epitope polymorphisms; epitope-HLA binding; lineages; protein ductility.
© The Author(s) 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
-
- Aranaz A, Cousins D, Mateos A, Dominguez L. 2003. Elevation of Mycobacterium tuberculosis subsp. caprae Aranaz et al. 1999 to species rank as Mycobacterium caprae comb. nov., sp. nov. Int J Syst Evol Microbiol. 53:1785–1789. - PubMed
-
- Babu MM, van der Lee R, de Groot NS, Gsponer J. 2011. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol. 21:432–440. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

