The OsMYB30 Transcription Factor Suppresses Cold Tolerance by Interacting with a JAZ Protein and Suppressing β-Amylase Expression
- PMID: 28062835
- PMCID: PMC5291022
- DOI: 10.1104/pp.16.01725
The OsMYB30 Transcription Factor Suppresses Cold Tolerance by Interacting with a JAZ Protein and Suppressing β-Amylase Expression
Abstract
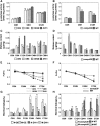
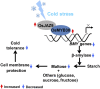
Cold stress is one of the major limiting factors for rice (Oryza sativa) productivity. Several MYB transcriptional factors have been reported as important regulators in the cold stress response, but the molecular mechanisms are largely unknown. In this study, we characterized a cold-responsive R2R3-type MYB gene, OsMYB30, for its regulatory function in cold tolerance in rice. Functional analysis revealed that overexpression of OsMYB30 in rice resulted in increased cold sensitivity, while the osmyb30 knockout mutant showed increased cold tolerance. Microarray and quantitative real-time polymerase chain reaction analyses revealed that a few β-amylase (BMY) genes were down-regulated by OsMYB30. The BMY activity and maltose content, which were decreased and increased in the OsMYB30 overexpression and osmyb30 knockout mutant, respectively, were correlated with the expression patterns of the BMY genes. OsMYB30 was shown to bind to the promoters of the BMY genes. These results suggested that OsMYB30 exhibited a regulatory effect on the breakdown of starch through the regulation of the BMY genes. In addition, application of maltose had a protective effect for cell membranes under cold stress conditions. Furthermore, we identified an OsMYB30-interacting protein, OsJAZ9, that had a significant effect in suppressing the transcriptional activation of OsMYB30 and in the repression of BMY genes mediated by OsMYB30. These results together suggested that OsMYB30 might be a novel regulator of cold tolerance through the negative regulation of the BMY genes by interacting with OsJAZ9 to fine-tune the starch breakdown and the content of maltose, which might contribute to the cold tolerance as a compatible solute.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures






References
-
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281: 37636–37645 - PubMed
-
- Bai Y, Meng Y, Huang D, Qi Y, Chen M (2011) Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98: 128–136 - PubMed
-
- Baker NR, Rosenqvist E (2004) Applications of chlorophyll fluorescence can improve crop production strategies: an examination of future possibilities. J Exp Bot 55: 1607–1621 - PubMed
-
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 - PubMed
-
- Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12: 444–451 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

