Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast
- PMID: 28062837
- PMCID: PMC5555582
- DOI: 10.1042/BCJ20160516
Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast
Abstract
Ribosome biogenesis requires the intertwined processes of folding, modification, and processing of ribosomal RNA, together with binding of ribosomal proteins. In eukaryotic cells, ribosome assembly begins in the nucleolus, continues in the nucleoplasm, and is not completed until after nascent particles are exported to the cytoplasm. The efficiency and fidelity of ribosome biogenesis are facilitated by >200 assembly factors and ∼76 different small nucleolar RNAs. The pathway is driven forward by numerous remodeling events to rearrange the ribonucleoprotein architecture of pre-ribosomes. Here, we describe principles of ribosome assembly that have emerged from recent studies of biogenesis of the large ribosomal subunit in the yeast Saccharomyces cerevisiae We describe tools that have empowered investigations of ribosome biogenesis, and then summarize recent discoveries about each of the consecutive steps of subunit assembly.
Keywords: ribonucleoproteins; ribosome assembly; ribosomes; yeast.
© 2017 The Author(s); published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
The Authors declare that there are no competing interests associated with the manuscript.
Figures




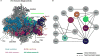
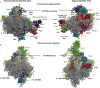
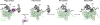
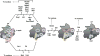
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

