Hijacking Complement Regulatory Proteins for Bacterial Immune Evasion
- PMID: 28066340
- PMCID: PMC5167704
- DOI: 10.3389/fmicb.2016.02004
Hijacking Complement Regulatory Proteins for Bacterial Immune Evasion
Abstract
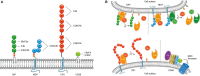
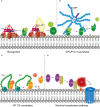
The human complement system plays an important role in the defense against invading pathogens, inflammation and homeostasis. Invading microbes, such as bacteria, directly activate the complement system resulting in the formation of chemoattractants and in effective labeling of the bacteria for phagocytosis. In addition, formation of the membrane attack complex is responsible for direct killing of Gram-negative bacteria. In turn, bacteria have evolved several ways to evade complement activation on their surface in order to be able to colonize and invade the human host. One important mechanism of bacterial escape is attraction of complement regulatory proteins to the microbial surface. These molecules are present in the human body for tight regulation of the complement system to prevent damage to host self-surfaces. Therefore, recruitment of complement regulatory proteins to the bacterial surface results in decreased complement activation on the microbial surface which favors bacterial survival. This review will discuss recent advances in understanding the binding of complement regulatory proteins to the bacterial surface at the molecular level. This includes, new insights that have become available concerning specific conserved motives on complement regulatory proteins that are favorable for microbial binding. Finally, complement evasion molecules are of high importance for vaccine development due to their dominant role in bacterial survival, high immunogenicity and homology as well as their presence on the bacterial surface. Here, the use of complement evasion molecules for vaccine development will be discussed.
Keywords: bacteria; complement; complement regulatory proteins; immune evasion of bacteria; vaccine development.
Figures




References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

