RXLR and CRN Effectors from the Sunflower Downy Mildew Pathogen Plasmopara halstedii Induce Hypersensitive-Like Responses in Resistant Sunflower Lines
- PMID: 28066456
- PMCID: PMC5165252
- DOI: 10.3389/fpls.2016.01887
RXLR and CRN Effectors from the Sunflower Downy Mildew Pathogen Plasmopara halstedii Induce Hypersensitive-Like Responses in Resistant Sunflower Lines
Abstract
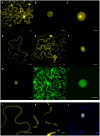
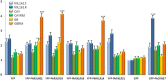
Plasmopara halstedii is an obligate biotrophic oomycete causing downy mildew disease on sunflower, Helianthus annuus, an economically important oil crop. Severe symptoms of the disease (e.g., plant dwarfism, leaf bleaching, sporulation and production of infertile flower) strongly impair seed yield. Pl resistance genes conferring resistance to specific P. halstedii pathotypes were located on sunflower genetic map but yet not cloned. They are present in cultivated lines to protect them against downy mildew disease. Among the 16 different P. halstedii pathotypes recorded in France, pathotype 710 is frequently found, and therefore continuously controlled in sunflower by different Pl genes. High-throughput sequencing of cDNA from P. halstedii led us to identify potential effectors with the characteristic RXLR or CRN motifs described in other oomycetes. Expression of six P. halstedii putative effectors, five RXLR and one CRN, was analyzed by qRT-PCR in pathogen spores and in the pathogen infecting sunflower leaves and selected for functional analyses. We developed a new method for transient expression in sunflower plant leaves and showed for the first time subcellular localization of P. halstedii effectors fused to a fluorescent protein in sunflower leaf cells. Overexpression of the CRN and of 3 RXLR effectors induced hypersensitive-like cell death reactions in some sunflower near-isogenic lines resistant to pathotype 710 and not in susceptible corresponding lines, suggesting they could be involved in Pl loci-mediated resistances.
Keywords: Agrobacterium-mediated transient expression; Plasmopara halstedii; downy mildew; hypersensitive response; oomycete effectors; resistant sunflower; subcellular localization.
Figures




References
-
- As-sadi F., Carrere S., Gascuel Q., Hourlier T., Rengel D., Le Paslier M.-C., et al. (2011). Transcriptomic analysis of the interaction between Helianthus annuus and its obligate parasite Plasmopara halstedii shows single nucleotide polymorphisms in CRN sequences. BMC Genomics 12:498 10.1186/1471-2164-12-498 - DOI - PMC - PubMed
-
- Bos J. I. B., Kanneganti T.-D., Young C., Cakir C., Huitema E., Win J., et al. (2006). The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48 165–176. 10.1111/j.1365-313X.2006.02866.x - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

