Coxiella burnetii Employs the Dot/Icm Type IV Secretion System to Modulate Host NF-κB/RelA Activation
- PMID: 28066723
- PMCID: PMC5165255
- DOI: 10.3389/fcimb.2016.00188
Coxiella burnetii Employs the Dot/Icm Type IV Secretion System to Modulate Host NF-κB/RelA Activation
Abstract
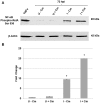
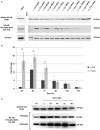
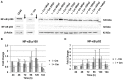
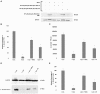
Coxiella burnetii is the causative agent of Q fever and an obligate intracellular pathogen in nature that survives and grows in a parasitophorous vacuole (PV) within eukaryotic host cells. C. burnetii promotes intracellular survival by subverting apoptotic and pro-inflammatory signaling pathways that are typically regulated by nuclear transcription factor-κB (NF-κB). We and others have demonstrated that C. burnetii NMII proteins inhibit expression of pro-inflammatory cytokines and induce expression of anti-apoptotic genes during infection. Here, we demonstrate that C. burnetii promotes intracellular survival by modulating NF-κB subunit p65 (RelA) phosphorylation, and thus activation, in a Type Four B Secretion System (T4BSS)-dependent manner. Immunoblot analysis of RelA phosphorylated at serine-536 demonstrated that C. burnetii increases NF-κB activation via the canonical pathway. However, RelA phosphorylation levels were even higher in infected cells where bacterial protein or mRNA synthesis was inhibited. Importantly, we demonstrate that inhibition of RelA phosphorylation impairs PV formation and C. burnetii growth. We found that a T4BSS-defective mutant (CbΔdotA) elicited phosphorylated RelA levels similar to those of wild type C. burnetii infection treated with Chloramphenicol. Moreover, cells infected with CbΔdotA or wild type C. burnetii treated with Chloramphenicol showed similar levels of GFP-RelA nuclear localization, and significantly increased localization compared to wild type C. burnetii infection. These data indicate that without de novo protein synthesis and a functional T4BSS, C. burnetii is unable to modulate NF-κB activation, which is crucial for optimal intracellular growth.
Keywords: Coxiella burnetii; NF-κB; Q fever; obligate intracellular; type four secretion system.
Figures
Similar articles
-
Coxiella burnetii Blocks Intracellular Interleukin-17 Signaling in Macrophages.Infect Immun. 2018 Sep 21;86(10):e00532-18. doi: 10.1128/IAI.00532-18. Print 2018 Oct. Infect Immun. 2018. PMID: 30061378 Free PMC article.
-
Dependency of Coxiella burnetii Type 4B Secretion on the Chaperone IcmS.J Bacteriol. 2019 Nov 5;201(23):e00431-19. doi: 10.1128/JB.00431-19. Print 2019 Dec 1. J Bacteriol. 2019. PMID: 31501284 Free PMC article.
-
Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages.mBio. 2011 Sep 1;2(4):e00175-11. doi: 10.1128/mBio.00175-11. Print 2011. mBio. 2011. PMID: 21862628 Free PMC article.
-
Coxiella burnetii as a useful tool to investigate bacteria-friendly host cell compartments.Int J Med Microbiol. 2018 Jan;308(1):77-83. doi: 10.1016/j.ijmm.2017.09.010. Epub 2017 Sep 14. Int J Med Microbiol. 2018. PMID: 28935173 Review.
-
Role of lipids in Coxiella burnetii infection.Adv Exp Med Biol. 2012;984:199-213. doi: 10.1007/978-94-007-4315-1_10. Adv Exp Med Biol. 2012. PMID: 22711633 Review.
Cited by
-
A CsrA-Binding, trans-Acting sRNA of Coxiella burnetii Is Necessary for Optimal Intracellular Growth and Vacuole Formation during Early Infection of Host Cells.J Bacteriol. 2019 Oct 21;201(22):e00524-19. doi: 10.1128/JB.00524-19. Print 2019 Nov 15. J Bacteriol. 2019. PMID: 31451541 Free PMC article.
-
Coxiella burnetii inhibits host immunity by a protein phosphatase adapted from glycolysis.Proc Natl Acad Sci U S A. 2022 Jan 4;119(1):e2110877119. doi: 10.1073/pnas.2110877119. Proc Natl Acad Sci U S A. 2022. PMID: 34930823 Free PMC article.
-
Rolling With Host Immunity: Virulence Beyond The Glycolysis.Front Cell Infect Microbiol. 2022 Jun 15;12:939828. doi: 10.3389/fcimb.2022.939828. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35782113 Free PMC article. No abstract available.
-
Coxiella burnetii Blocks Intracellular Interleukin-17 Signaling in Macrophages.Infect Immun. 2018 Sep 21;86(10):e00532-18. doi: 10.1128/IAI.00532-18. Print 2018 Oct. Infect Immun. 2018. PMID: 30061378 Free PMC article.
-
CYP1B1-AS1 regulates CYP1B1 to promote Coxiella burnetii pathogenesis by inhibiting ROS and host cell death.Res Sq [Preprint]. 2024 Dec 11:rs.3.rs-5390645. doi: 10.21203/rs.3.rs-5390645/v1. Res Sq. 2024. PMID: 39711545 Free PMC article. Preprint.
References
-
- Amara A. B., Bechah Y., Mege J.-L. (2012). Immune response and Coxiella burnetii invasion, in Coxiella burnetii: Recent Advances and New Perspectives in Research of the Q Fever Bacterium, eds Toman R., Heinzen A. R., Samuel E. J., Mege J.-L. (Dordrecht: Springer Netherlands; ), 287–298. - PubMed
-
- Andoh M., Zhang G., Russell-Lodrigue K. E., Shive H. R., Weeks B. R., Samuel J. E. (2007). T cells are essential for bacterial clearance, and gamma interferon, tumor necrosis factor alpha, and B cells are crucial for disease development in Coxiella burnetii infection in mice. Infect. Immun. 75, 3245–3255. 10.1128/IAI.01767-06 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

