Analyzing native membrane protein assembly in nanodiscs by combined non-covalent mass spectrometry and synthetic biology
- PMID: 28067619
- PMCID: PMC5291076
- DOI: 10.7554/eLife.20954
Analyzing native membrane protein assembly in nanodiscs by combined non-covalent mass spectrometry and synthetic biology
Abstract
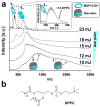
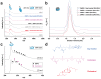
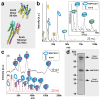
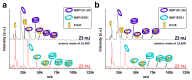
Membrane proteins frequently assemble into higher order homo- or hetero-oligomers within their natural lipid environment. This complex formation can modulate their folding, activity as well as substrate selectivity. Non-disruptive methods avoiding critical steps, such as membrane disintegration, transfer into artificial environments or chemical modifications are therefore essential to analyze molecular mechanisms of native membrane protein assemblies. The combination of cell-free synthetic biology, nanodisc-technology and non-covalent mass spectrometry provides excellent synergies for the analysis of membrane protein oligomerization within defined membranes. We exemplify our strategy by oligomeric state characterization of various membrane proteins including ion channels, transporters and membrane-integrated enzymes assembling up to hexameric complexes. We further indicate a lipid-dependent dimer formation of MraY translocase correlating with the enzymatic activity. The detergent-free synthesis of membrane protein/nanodisc samples and the analysis by LILBID mass spectrometry provide a versatile platform for the analysis of membrane proteins in a native environment.
Keywords: biochemistry; biophysics; cell-free synthetic biology; membrane protein; nanodisc; non-covalent mass spectrometry; none; oligomeric state; structural biology.
Conflict of interest statement
VD: Reviewing editor, eLife. The other authors declare that no competing interests exist.
Figures








References
-
- Bugg TD, Rodolis MT, Mihalyi A, Jamshidi S. Inhibition of phospho-MurNAc-pentapeptide translocase (MraY) by nucleoside natural product antibiotics, bacteriophage ϕX174 lysis protein E, and cationic antibacterial peptides. Bioorganic & Medicinal Chemistry. 2016;24:6340–6347. doi: 10.1016/j.bmc.2016.03.018. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

