Effects of Replication and Transcription on DNA Structure-Related Genetic Instability
- PMID: 28067787
- PMCID: PMC5295012
- DOI: 10.3390/genes8010017
Effects of Replication and Transcription on DNA Structure-Related Genetic Instability
Abstract
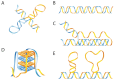
Many repetitive sequences in the human genome can adopt conformations that differ from the canonical B-DNA double helix (i.e., non-B DNA), and can impact important biological processes such as DNA replication, transcription, recombination, telomere maintenance, viral integration, transposome activation, DNA damage and repair. Thus, non-B DNA-forming sequences have been implicated in genetic instability and disease development. In this article, we discuss the interactions of non-B DNA with the replication and/or transcription machinery, particularly in disease states (e.g., tumors) that can lead to an abnormal cellular environment, and how such interactions may alter DNA replication and transcription, leading to potential conflicts at non-B DNA regions, and eventually result in genetic stability and human disease.
Keywords: DNA repair; collision; genetic instability; non-B DNA; replication; transcription.
Conflict of interest statement
The authors declare no conflict of interest. The funding sponsor had no role in the writing of the review manuscript.
Figures


References
-
- Chen X., Shen Y., Zhang F., Chiang C., Pillalamarri V., Blumenthal I., Talkowski M., Wu B.L., Gusella J.F. Molecular analysis of a deletion hotspot in the NRXN1 region reveals the involvement of short inverted repeats in deletion CNVs. Am. J. Hum. Genet. 2013;92:375–386. doi: 10.1016/j.ajhg.2013.02.006. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

