Social and Genetic Networks of HIV-1 Transmission in New York City
- PMID: 28068413
- PMCID: PMC5221827
- DOI: 10.1371/journal.ppat.1006000
Social and Genetic Networks of HIV-1 Transmission in New York City
Abstract
Background: Sexually transmitted infections spread across contact networks. Partner elicitation and notification are commonly used public health tools to identify, notify, and offer testing to persons linked in these contact networks. For HIV-1, a rapidly evolving pathogen with low per-contact transmission rates, viral genetic sequences are an additional source of data that can be used to infer or refine transmission networks.
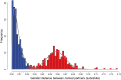
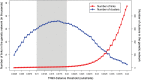
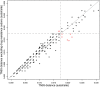
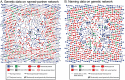
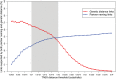
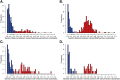
Methods and findings: The New York City Department of Health and Mental Hygiene interviews individuals newly diagnosed with HIV and elicits names of sexual and injection drug using partners. By law, the Department of Health also receives HIV sequences when these individuals enter healthcare and their physicians order resistance testing. Our study used both HIV sequence and partner naming data from 1342 HIV-infected persons in New York City between 2006 and 2012 to infer and compare sexual/drug-use named partner and genetic transmission networks. Using these networks, we determined a range of genetic distance thresholds suitable for identifying potential transmission partners. In 48% of cases, named partners were infected with genetically closely related viruses, compatible with but not necessarily representing or implying, direct transmission. Partner pairs linked through the genetic similarity of their HIV sequences were also linked by naming in 53% of cases. Persons who reported high-risk heterosexual contact were more likely to name at least one partner with a genetically similar virus than those reporting their risk as injection drug use or men who have sex with men.
Conclusions: We analyzed an unprecedentedly large and detailed partner tracing and HIV sequence dataset and determined an empirically justified range of genetic distance thresholds for identifying potential transmission partners. We conclude that genetic linkage provides more reliable evidence for identifying potential transmission partners than partner naming, highlighting the importance and complementarity of both epidemiological and molecular genetic surveillance for characterizing regional HIV-1 epidemics.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: JOW is a paid consultant for the Centers for Disease Control and Prevention.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

