Complex Subtype Diversity of HIV-1 Among Drug Users in Major Kenyan Cities
- PMID: 28068781
- PMCID: PMC5439455
- DOI: 10.1089/AID.2016.0321
Complex Subtype Diversity of HIV-1 Among Drug Users in Major Kenyan Cities
Abstract
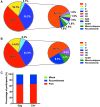
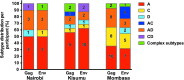
Drug users are increasingly recognized as a key population driving human immunodeficiency virus (HIV) spread in sub-Saharan Africa. To determine HIV-1 subtypes circulating in this population group and explore possible geographic differences, we analyzed HIV-1 sequences among drug users from Nairobi, Mombasa, and Kisumu in Kenya. We sequenced gag and env from 55 drug users. Subtype analysis from 220 gag clonal sequences from 54 of 55 participants (median = 4/participant) showed that 44.4% were A, 16.7% were C, 3.7% were D, and 35.2% were intersubtype recombinants. Of 156 env clonal sequences from 48 of 55 subjects (median = 3/participant), 45.8% were subtype A, 14.6% were C, 6.3% were D, and 33.3% were recombinants. Comparative analysis of both genes showed that 30 (63.8%) participants had concordant subtypes, while 17 (36.2%) were discordant. We identified one genetically linked transmission pair and two cases of dual infection. These data are indicative of extensive HIV-1 intersubtype recombination in Kenya and suggest decline in subtype D prevalence.
Keywords: HIV-1 subtypes; Kenya; drug users; intersubtype recombinants.
Conflict of interest statement
No competing financial interests exist.
Figures





References
-
- Virgin HW, Walker BD: Immunology and the elusive AIDS vaccine. Nature 2010;464:224–231 - PubMed
-
- Ndung'u T, Weiss RA: On HIV diversity. AIDS 2012;26:1255–1260 - PubMed
-
- Osman S, Lihana RW, Kibaya RM, et al. : Diversity of HIV type 1 and drug resistance mutations among injecting drug users in Kenya. AIDS Res Hum Retroviruses 2013;29:187–190 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

