Domestication of rice has reduced the occurrence of transposable elements within gene coding regions
- PMID: 28068923
- PMCID: PMC5223533
- DOI: 10.1186/s12864-016-3454-z
Domestication of rice has reduced the occurrence of transposable elements within gene coding regions
Abstract
Background: Transposable elements (TEs) are prominent features in many plant genomes, and patterns of TEs in closely related rice species are thus proposed as an ideal model to study TEs roles in the context of plant genome evolution. As TEs may contribute to improved rice growth and grain quality, it is of pivotal significance for worldwide food security and biomass production.
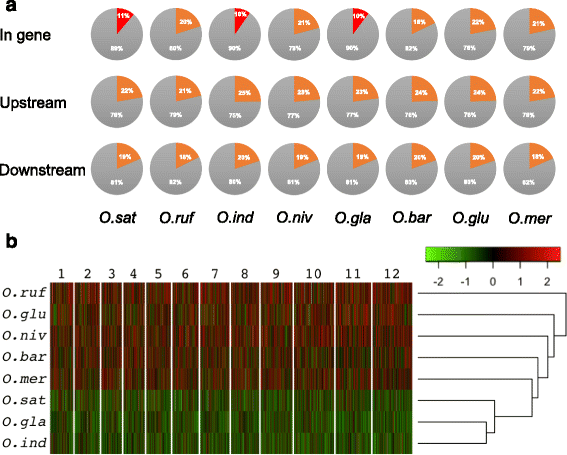
Results: We analyzed three cultivated rice species and their closest five wild relatives for distribution and content of TEs in their genomes. Despite that the three cultivar rice species contained similar copies and more total TEs, their genomes contained much longer TEs as compared to their wild relatives. Notably, TEs were largely depleted from genomic regions that corresponded to genes in the cultivated species, while this was not the case for their wild relatives. Gene ontology and gene homology analyses revealed that while certain genes contained TEs in all the wild species, the closest homologs in the cultivated species were devoid of them. This distribution of TEs is surprising as the cultivated species are more distantly related to each other as compared to their closest wild relative. Hence, cultivated rice species have more similar TE distributions among their genes as compared to their closest wild relatives. We, furthermore, exemplify how genes that are conferring important rice traits can be regulated by TE associations.
Conclusions: This study demonstrate that the cultivation of rice has led to distinct genomic distribution of TEs, and that certain rice traits are closely associated with TE distribution patterns. Hence, the results provide means to better understand TE-dependent rice traits and the potential to genetically engineer rice for better performance.
Keywords: Cultivated rice; Evolution; Oryza; Transposable elements; Wild rice.
Figures






Similar articles
-
Genome-wide analysis of Dongxiang wild rice (Oryza rufipogon Griff.) to investigate lost/acquired genes during rice domestication.BMC Plant Biol. 2016 Apr 26;16:103. doi: 10.1186/s12870-016-0788-2. BMC Plant Biol. 2016. PMID: 27118394 Free PMC article.
-
Rice structural variation: a comparative analysis of structural variation between rice and three of its closest relatives in the genus Oryza.Plant J. 2010 Sep;63(6):990-1003. doi: 10.1111/j.1365-313X.2010.04293.x. Plant J. 2010. PMID: 20626650
-
Rice transposable elements are characterized by various methylation environments in the genome.BMC Genomics. 2007 Dec 20;8:469. doi: 10.1186/1471-2164-8-469. BMC Genomics. 2007. PMID: 18093338 Free PMC article.
-
What makes up plant genomes: The vanishing line between transposable elements and genes.Biochim Biophys Acta. 2016 Feb;1859(2):366-80. doi: 10.1016/j.bbagrm.2015.12.005. Epub 2015 Dec 17. Biochim Biophys Acta. 2016. PMID: 26709091 Review.
-
Transposable element influences on gene expression in plants.Biochim Biophys Acta Gene Regul Mech. 2017 Jan;1860(1):157-165. doi: 10.1016/j.bbagrm.2016.05.010. Epub 2016 May 25. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 27235540 Review.
Cited by
-
Transcriptional regulation of genes bearing intronic heterochromatin in the rice genome.PLoS Genet. 2020 Mar 18;16(3):e1008637. doi: 10.1371/journal.pgen.1008637. eCollection 2020 Mar. PLoS Genet. 2020. PMID: 32187179 Free PMC article.
-
Miniature Inverted-Repeat Transposable Elements: Small DNA Transposons That Have Contributed to Plant MICRORNA Gene Evolution.Plants (Basel). 2023 Mar 1;12(5):1101. doi: 10.3390/plants12051101. Plants (Basel). 2023. PMID: 36903960 Free PMC article. Review.
-
Development of molecular markers based on LTR retrotransposon in the Cleistogenes songorica genome.J Appl Genet. 2022 Feb;63(1):61-72. doi: 10.1007/s13353-021-00658-9. Epub 2021 Sep 23. J Appl Genet. 2022. PMID: 34554437
-
Chromosome-Scale Assembly of Winter Oilseed Rape Brassica napus.Front Plant Sci. 2020 Apr 28;11:496. doi: 10.3389/fpls.2020.00496. eCollection 2020. Front Plant Sci. 2020. PMID: 32411167 Free PMC article.
-
New clues into the mechanisms of rice domestication.J Biosci. 2019 Jun;44(2):28. J Biosci. 2019. PMID: 31180041 Review.
References
-
- Wessler SR, Nagel A, Casa A. Miniature inverted repeat transposable elements help create genomic diversity in maize and rice. In: Khush GS, Brar DS, Hardy B, editors. Rice genetics IV proceedings of the fourth international rice genetics symposium. Los Baños: Philippines; 2001. pp. 107–16.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

