The Influence of the Microbiome on Allergic Sensitization to Food
- PMID: 28069753
- PMCID: PMC5228404
- DOI: 10.4049/jimmunol.1601266
The Influence of the Microbiome on Allergic Sensitization to Food
Abstract
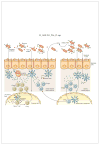
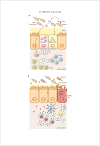
The alarming increase in the incidence and severity of food allergies has coincided with lifestyle changes in Western societies, such as dietary modifications and increased antibiotic use. These demographic shifts have profoundly altered the coevolved relationship between host and microbiota, depleting bacterial populations critical for the maintenance of mucosal homeostasis. There is increasing evidence that the dysbiosis associated with sensitization to food fails to stimulate protective tolerogenic pathways, leading to the development of the type 2 immune responses that characterize allergic disease. Defining the role of beneficial allergy-protective members of the microbiota in the regulation of tolerance to food has exciting potential for new interventions to treat dietary allergies by modulation of the microbiota.
Copyright © 2017 by The American Association of Immunologists, Inc.
Figures
References
-
- Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, Jansson JK, Dorrestein PC, Knight R. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature. 2016;535:94–103. - PubMed
-
- Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

