Residues in the RecQ C-terminal Domain of the Human Werner Syndrome Helicase Are Involved in Unwinding G-quadruplex DNA
- PMID: 28069813
- PMCID: PMC5336152
- DOI: 10.1074/jbc.M116.767699
Residues in the RecQ C-terminal Domain of the Human Werner Syndrome Helicase Are Involved in Unwinding G-quadruplex DNA
Abstract
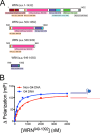
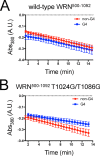
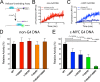
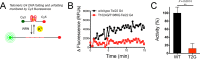
The structural and biophysical properties typically associated with G-quadruplex (G4) structures render them a significant block for DNA replication, which must be overcome for cell division to occur. The Werner syndrome protein (WRN) is a RecQ family helicase that has been implicated in the efficient processing of G4 DNA structures. The aim of this study was to identify the residues of WRN involved in the binding and ATPase-driven unwinding of G4 DNA. Using a c-Myc G4 DNA model sequence and recombinant WRN, we have determined that the RecQ-C-terminal (RQC) domain of WRN imparts a 2-fold preference for binding to G4 DNA relative to non-G4 DNA substrates. NMR studies identified residues involved specifically in interactions with G4 DNA. Three of the amino acids in the WRN RQC domain that exhibited the largest G4-specific changes in NMR signal were then mutated alone or in combination. Mutating individual residues implicated in G4 binding had a modest effect on WRN binding to DNA, decreasing the preference for G4 substrates by ∼25%. Mutating two G4-interacting residues (T1024G and T1086G) abrogated preferential binding of WRN to G4 DNA. Very modest decreases in G4 DNA-stimulated ATPase activity were observed for the mutant enzymes. Most strikingly, G4 unwinding by WRN was inhibited ∼50% for all three point mutants and >90% for the WRN double mutant (T1024G/T1086G) relative to normal B-form dsDNA substrates. Our work has helped to identify residues in the WRN RQC domain that are involved specifically in the interaction with G4 DNA.
Keywords: DNA helicase; DNA repair; DNA replication; G-quadruplex; Werner syndrome helicase; enzyme mechanism.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

References
-
- Delagoutte E., and von Hippel P. H. (2002) Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: structures and properties of isolated helicases. Q. Rev. Biophys. 35, 431–478 - PubMed
-
- Eoff R. L., and Raney K. D. (2005) Helicase-catalysed translocation and strand separation. Biochem. Soc. Trans. 33, 1474–1478 - PubMed
-
- Wu C. G., and Spies M. (2013) Overview: what are helicases? Adv. Exp. Med. Biol. 767, 1–16 - PubMed
-
- Caruthers J. M., and McKay D. B. (2002) Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12, 123–133 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

