Quality control mechanisms exclude incorrect polymerases from the eukaryotic replication fork
- PMID: 28069954
- PMCID: PMC5278475
- DOI: 10.1073/pnas.1619748114
Quality control mechanisms exclude incorrect polymerases from the eukaryotic replication fork
Abstract
The eukaryotic genome is primarily replicated by two DNA polymerases, Pol ε and Pol δ, that function on the leading and lagging strands, respectively. Previous studies have established recruitment mechanisms whereby Cdc45-Mcm2-7-GINS (CMG) helicase binds Pol ε and tethers it to the leading strand, and PCNA (proliferating cell nuclear antigen) binds tightly to Pol δ and recruits it to the lagging strand. The current report identifies quality control mechanisms that exclude the improper polymerase from a particular strand. We find that the replication factor C (RFC) clamp loader specifically inhibits Pol ε on the lagging strand, and CMG protects Pol ε against RFC inhibition on the leading strand. Previous studies show that Pol δ is slow and distributive with CMG on the leading strand. However, Saccharomyces cerevisiae Pol δ-PCNA is a rapid and processive enzyme, suggesting that CMG may bind and alter Pol δ activity or position it on the lagging strand. Measurements of polymerase binding to CMG demonstrate Pol ε binds CMG with a Kd value of 12 nM, but Pol δ binding CMG is undetectable. Pol δ, like bacterial replicases, undergoes collision release upon completing replication, and we propose Pol δ-PCNA collides with the slower CMG, and in the absence of a stabilizing Pol δ-CMG interaction, the collision release process is triggered, ejecting Pol δ on the leading strand. Hence, by eviction of incorrect polymerases at the fork, the clamp machinery directs quality control on the lagging strand and CMG enforces quality control on the leading strand.
Keywords: DNA polymerase; PCNA; clamp loader; replication; replisome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









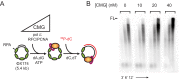





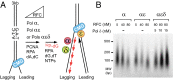

References
-
- Kornberg A, Baker TA. DNA Replication. 2nd Ed University Science Books; Herndon, VA: 2005.
-
- Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70(1):181–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

