Noncontiguous finished genome sequence and description of Prevotella phocaeensis sp. nov., a new anaerobic species isolated from human gut infected by Clostridium difficile
- PMID: 28070336
- PMCID: PMC5219628
- DOI: 10.1016/j.nmni.2016.12.013
Noncontiguous finished genome sequence and description of Prevotella phocaeensis sp. nov., a new anaerobic species isolated from human gut infected by Clostridium difficile
Abstract
Prevotella phocaeensis sp. nov. strain SN19T (= DSM 103364) is a new species isolated from the gut microbiota of patient with colitis due to Clostridium difficile. Strain SN19T is Gram-negative rod-shaped bacteria, strictly anaerobic, nonmotile and non-endospore forming. The predominance fatty acid is hexadecanoic acid. Its 16S rRNA showed a 97.70% sequence identity with its phylogenetically closest species, Prevotella oralis. The genome is 2 922 117 bp long and contains 2486 predicted genes including 56 RNA genes.
Keywords: Clostridium difficile; Prevotella phocaeensis; culturomics; gut microbiota; taxonogenomics.
Figures
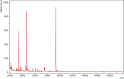




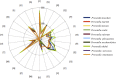
References
-
- Landman C., Quévrain E. Le microbiote intestinal: description, rôle et implications physiopathologiques. Rev Med Interne. 2016;37:418–423. - PubMed
-
- Hooper L.V. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12:129–134. - PubMed
-
- Christl S.U., Murgatroyd P.R., Gibson G.R., Cummings J.H. Production, metabolism, and excretion of hydrogen in the large intestine. Gastroenterology. 1992;102:1269–1277. - PubMed
-
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
