Gut Microbiome of the Canadian Arctic Inuit
- PMID: 28070563
- PMCID: PMC5214747
- DOI: 10.1128/mSphere.00297-16
Gut Microbiome of the Canadian Arctic Inuit
Abstract
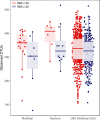
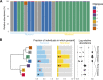
Diet is a major determinant of community composition in the human gut microbiome, and "traditional" diets have been associated with distinct and highly diverse communities, compared to Western diets. However, most traditional diets studied have been those of agrarians and hunter-gatherers consuming fiber-rich diets. In contrast, the Inuit of the Canadian Arctic have been consuming a traditional diet low in carbohydrates and rich in animal fats and protein for thousands of years. We hypothesized that the Inuit diet and lifestyle would be associated with a distinct microbiome. We used deep sequencing of the 16S rRNA gene to compare the gut microbiomes of Montrealers with a Western diet to those of the Inuit consuming a range of traditional and Western diets. At the overall microbial community level, the gut microbiomes of Montrealers and Inuit were indistinguishable and contained similar levels of microbial diversity. However, we observed significant differences in the relative abundances of certain microbial taxa down to the subgenus level using oligotyping. For example, Prevotella spp., which have been previously associated with high-fiber diets, were enriched in Montrealers and among the Inuit consuming a Western diet. The gut microbiomes of Inuit consuming a traditional diet also had significantly less genetic diversity within the Prevotella genus, suggesting that a low-fiber diet might not only select against Prevotella but also reduce its diversity. Other microbes, such as Akkermansia, were associated with geography as well as diet, suggesting limited dispersal to the Arctic. Our report provides a snapshot of the Inuit microbiome as Western-like in overall community structure but distinct in the relative abundances and diversity of certain genera and strains. IMPORTANCE Non-Western populations have been shown to have distinct gut microbial communities shaped by traditional diets. The hitherto-uncharacterized microbiome of the Inuit may help us to better understand health risks specific to this population such as diabetes and obesity, which increase in prevalence as many Inuit transition to a Western diet. Here we show that even Inuit consuming a mostly traditional diet have a broadly Western-like microbiome. This suggests that similarities between the Inuit diet and the Western diet (low fiber, high fat) may lead to a convergence of community structures and diversity. However, certain species and strains of microbes have significantly different levels of abundance and diversity in the Inuit, possibly driven by differences in diet. Furthermore, the Inuit diet provides an exception to the correlation between traditional diets and high microbial diversity, potentially due to their transitioning diet. Knowledge of the Inuit microbiome may provide future resources for interventions and conservation of Inuit heritage.
Keywords: Akkermansia; Inuit microbiome; Prevotella; Western diet; alpha diversity; oligotyping; traditional diet.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
