Dynamic relationships between ribosomal conformational and RNA positional changes during ribosomal translocation
- PMID: 28070564
- PMCID: PMC5219732
- DOI: 10.1016/j.heliyon.2016.e00214
Dynamic relationships between ribosomal conformational and RNA positional changes during ribosomal translocation
Abstract
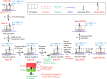
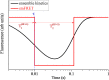
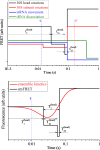
Ribosomal translocation catalyzed by EF-G hydrolyzing GTP entails multiple conformational changes of ribosome and positional changes of tRNAs and mRNA in the ribosome. However, the detailed dynamic relations among these changes and EF-G sampling are not clear. Here, based on our proposed pathway of ribosomal translocation, we study theoretically the dynamic relations among these changes exhibited in the single molecule data and those exhibited in the ensemble kinetic data. It is shown that the timing of these changes in the single molecule data and that in the ensemble kinetic data show very different. The theoretical results are in agreement with both the available ensemble kinetic experimental data and the single molecule experimental data.
Keywords: Biophysics; Mathematical Bioscience.
Figures


Similar articles
-
On the pathway of ribosomal translocation.Int J Biol Macromol. 2016 Nov;92:401-415. doi: 10.1016/j.ijbiomac.2016.07.048. Epub 2016 Jul 16. Int J Biol Macromol. 2016. PMID: 27431796
-
[Mechanism of tRNA translocation on the ribosome].Mol Biol (Mosk). 2001 Jul-Aug;35(4):655-65. Mol Biol (Mosk). 2001. PMID: 11524952 Review. Russian.
-
Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation.J Mol Biol. 2004 Nov 5;343(5):1183-94. doi: 10.1016/j.jmb.2004.08.097. J Mol Biol. 2004. PMID: 15491605
-
Spontaneous ribosomal translocation of mRNA and tRNAs into a chimeric hybrid state.Proc Natl Acad Sci U S A. 2019 Apr 16;116(16):7813-7818. doi: 10.1073/pnas.1901310116. Epub 2019 Apr 1. Proc Natl Acad Sci U S A. 2019. PMID: 30936299 Free PMC article.
-
The ribosome as a molecular machine: the mechanism of tRNA-mRNA movement in translocation.Biochem Soc Trans. 2011 Apr;39(2):658-62. doi: 10.1042/BST0390658. Biochem Soc Trans. 2011. PMID: 21428957 Review.
Cited by
-
Processivity of dimeric kinesin-1 molecular motors.FEBS Open Bio. 2018 Jul 20;8(8):1332-1351. doi: 10.1002/2211-5463.12486. eCollection 2018 Aug. FEBS Open Bio. 2018. PMID: 30087836 Free PMC article.
-
Mechanism of ribosome translation through mRNA secondary structures.Int J Biol Sci. 2017 May 16;13(6):712-722. doi: 10.7150/ijbs.19508. eCollection 2017. Int J Biol Sci. 2017. PMID: 28655997 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

