p-Cymene Promotes Its Catabolism through the p-Cymene and the p-Cumate Pathways, Activates a Stress Response and Reduces the Biofilm Formation in Burkholderia xenovorans LB400
- PMID: 28072820
- PMCID: PMC5224996
- DOI: 10.1371/journal.pone.0169544
p-Cymene Promotes Its Catabolism through the p-Cymene and the p-Cumate Pathways, Activates a Stress Response and Reduces the Biofilm Formation in Burkholderia xenovorans LB400
Abstract
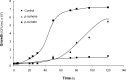
p-Cymene is an aromatic terpene that is present in diverse plant species. The aims of this study were to study the p-cymene metabolism in the model aromatic-degrading bacterium Burkholderia xenovorans LB400, and its response to p-cymene. The catabolic p-cymene (cym) and p-cumate (cmt) genes are clustered on the LB400 major chromosome. B. xenovorans LB400 was able to grow on p-cymene as well as on p-cumate as a sole carbon and energy sources. LB400 growth attained higher cell concentration at stationary phase on p-cumate than on p-cymene. The transcription of the key cymAb and cmtAb genes, and p-cumate dioxygenase activity were observed in LB400 cells grown on p-cymene and on p-cumate, but not in glucose-grown cells. Diverse changes on LB400 proteome were observed in p-cymene-grown cells compared to glucose-grown cells. An increase of the molecular chaperones DnaK, GroEL and ClpB, the organic hydroperoxide resistance protein Ohr, the alkyl hydroperoxide reductase AhpC and the copper oxidase CopA during growth on p-cymene strongly suggests that the exposure to p-cymene constitutes a stress condition for strain LB400. Diverse proteins of the energy metabolism such as enolase, pyruvate kinase, aconitase AcnA, succinyl-CoA synthetase beta subunit and ATP synthase beta subunit were induced by p-cymene. Electron microscopy showed that p-cymene-grown cells exhibited fuzzy outer and inner membranes and an increased periplasm. p-Cymene induced diverse membrane and transport proteins including the p-cymene transporter CymD. Biofilm formation was reduced during growth in p-cymene in strain LB400 compared to glucose-grown cells that may be associated with a decrease of diguanylate cyclase protein levels. Overall, these results indicate active p-cymene and p-cumate catabolic pathways in B. xenovorans LB400. In addition, this study showed that p-cymene activated a stress response in strain LB400 and reduced its biofilm formation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- Radke M. (1987) Organic geochemistry of aromatic hydrocarbons. Adv Petrol Geochem 2:141–207.
-
- Gildemeister E, Hoffman F. (1960). “Die ätherischen Öle”Vol. 4, Akademie-Verlag, Berlin.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

