Epigenetic siRNA and Chemical Screens Identify SETD8 Inhibition as a Therapeutic Strategy for p53 Activation in High-Risk Neuroblastoma
- PMID: 28073004
- PMCID: PMC5233415
- DOI: 10.1016/j.ccell.2016.12.002
Epigenetic siRNA and Chemical Screens Identify SETD8 Inhibition as a Therapeutic Strategy for p53 Activation in High-Risk Neuroblastoma
Abstract
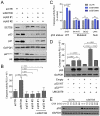
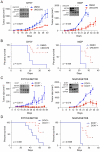
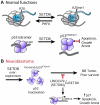
Given the paucity of druggable mutations in high-risk neuroblastoma (NB), we undertook chromatin-focused small interfering RNA and chemical screens to uncover epigenetic regulators critical for the differentiation block in high-risk NB. High-content Opera imaging identified 53 genes whose loss of expression led to a decrease in NB cell proliferation and 16 also induced differentiation. From these, the secondary chemical screen identified SETD8, the H4K20me1 methyltransferase, as a druggable NB target. Functional studies revealed that SETD8 ablation rescued the pro-apoptotic and cell-cycle arrest functions of p53 by decreasing p53K382me1, leading to activation of the p53 canonical pathway. In pre-clinical xenograft NB models, genetic or pharmacological (UNC0379) SETD8 inhibition conferred a significant survival advantage, providing evidence for SETD8 as a therapeutic target in NB.
Keywords: SETD8; differentiation; epigenetics; neuroblastoma; p53; siRNA screen.
Published by Elsevier Inc.
Figures





References
-
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. - PubMed
-
- Bagatell R, Beck-Popovic M, London WB, Zhang Y, Pearson AD, Matthay KK, Monclair T, Ambros PF, Cohn SL. Significance of MYCN amplification in international neuroblastoma staging system stage 1 and 2 neuroblastoma: a report from the International Neuroblastoma Risk Group database. J Clin Oncol. 2009;27:365–370. - PMC - PubMed
-
- Barbieri E, De Preter K, Capasso M, Chen Z, Hsu DM, Tonini GP, Lefever S, Hicks J, Versteeg R, Pession A, et al. Histone chaperone CHAF1A inhibits differentiation and promotes aggressive neuroblastoma. Cancer Res. 2014;74:765–774. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

