Evolution of condensin and cohesin complexes driven by replacement of Kite by Hawk proteins
- PMID: 28073014
- PMCID: PMC5228436
- DOI: 10.1016/j.cub.2016.11.050
Evolution of condensin and cohesin complexes driven by replacement of Kite by Hawk proteins
Abstract
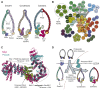
Mitotic chromosome condensation, sister chromatid cohesion, and higher order folding of interphase chromatin are mediated by condensin and cohesin, eukaryotic members of the SMC (structural maintenance of chromosomes)-kleisin protein family. Other members facilitate chromosome segregation in bacteria [1]. A hallmark of these complexes is the binding of the two ends of a kleisin subunit to the apices of V-shaped Smc dimers, creating a tripartite ring capable of entrapping DNA (Figure 1A). In addition to creating rings, kleisins recruit regulatory subunits. One family of regulators, namely Kite dimers (Kleisin interacting winged-helix tandem elements), interact with Smc-kleisin rings from bacteria, archaea and the eukaryotic Smc5-6 complex, but not with either condensin or cohesin [2]. These instead possess proteins containing HEAT (Huntingtin/EF3/PP2A/Tor1) repeat domains whose origin and distribution have not yet been characterized. Using a combination of profile Hidden Markov Model (HMM)-based homology searches, network analysis and structural alignments, we identify a common origin for these regulators, for which we propose the name Hawks, i.e. HEAT proteins associated with kleisins.
Copyright © 2017 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
References
-
- Nasmyth K., Haering C.H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. - PubMed
-
- Palecek J.J., Gruber S. Kite proteins: a superfamily of SMC/Kleisin partners conserved across Bacteria, Archaea, and Eukaryotes. Structure. 2015;23:2183–2190. - PubMed
-
- Andrade M.A., Petosa C., O’Donoghue S.I., Müller C.W., Bork P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. - PubMed
-
- Remmert M., Biegert A., Hauser A., Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 2011;9:173–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

