Transcription Factors PvERF15 and PvMTF-1 Form a Cadmium Stress Transcriptional Pathway
- PMID: 28073984
- PMCID: PMC5338663
- DOI: 10.1104/pp.16.01729
Transcription Factors PvERF15 and PvMTF-1 Form a Cadmium Stress Transcriptional Pathway
Abstract
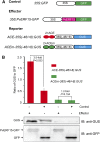
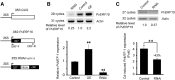
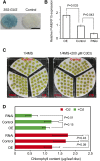
In plants, cadmium (Cd)-responsive transcription factors are key downstream effectors of Cd stress transcriptional pathways, which are capable of converging Cd stress signals through triggering the expression of Cd detoxification genes. However, the upstream transcriptional regulatory pathways that modulate their responses to Cd are less clear. Previously, we identified the bean (Phaseolus vulgaris) METAL RESPONSE ELEMENT-BINDING TRANSCRIPTION FACTOR1 (PvMTF-1) that responds to Cd and confers Cd tolerance in planta. Here, we demonstrate an upstream transcriptional regulation of the PvMTF-1 response to Cd Using a yeast one-hybrid system, we cloned the bean ETHYLENE RESPONSE FACTOR15 (PvERF15) that binds to the PvMTF-1 promoter. PvERF15 was strongly induced by Cd stress, and its overexpression resulted in the up-regulation of PvMTF-1 DNA-protein interaction assays further revealed that PvERF15 binds directly to a 19-bp AC-rich element in the PvMTF-1 promoter. The AC-rich element serves as a positive element bound by PvERF15 to activate gene expression. More importantly, knockdown of PvERF15 by RNA interference resulted in reduced Cd-induced expression of PvMTF-1PvERF15 seems to be involved in Cd tolerance, since knockdown of PvERF15 by RNA interference in bean leaf discs decreased Cd tolerance in a transient assay. Since PvERF15 is a component of the Cd stress transcriptional pathway in beans and PvMTF-1 is one of its downstream targets, our findings provide a PvERF15/PvMTF-1 transcriptional pathway and thereby contribute to the understanding of Cd stress transcriptional regulatory pathways in plants.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures



Similar articles
-
An AC-Rich Bean Element Serves as an Ethylene-Responsive Element in Arabidopsis.Plants (Basel). 2020 Aug 14;9(8):1033. doi: 10.3390/plants9081033. Plants (Basel). 2020. PMID: 32823972 Free PMC article.
-
Bean metal-responsive element-binding transcription factor confers cadmium resistance in tobacco.Plant Physiol. 2015 Mar;167(3):1136-48. doi: 10.1104/pp.114.253096. Epub 2015 Jan 26. Plant Physiol. 2015. PMID: 25624396 Free PMC article.
-
Intronic promoter-mediated feedback loop regulates bean PvSR2 gene expression.Biochem Biophys Res Commun. 2015 Aug 7;463(4):1097-101. doi: 10.1016/j.bbrc.2015.06.064. Epub 2015 Jun 12. Biochem Biophys Res Commun. 2015. PMID: 26079876
-
Metal transport proteins and transcription factor networks in plant responses to cadmium stress.Plant Cell Rep. 2024 Aug 17;43(9):218. doi: 10.1007/s00299-024-03303-x. Plant Cell Rep. 2024. PMID: 39153039 Review.
-
Transcriptional Regulatory Network of Plant Cadmium Stress Response.Int J Mol Sci. 2023 Feb 22;24(5):4378. doi: 10.3390/ijms24054378. Int J Mol Sci. 2023. PMID: 36901809 Free PMC article. Review.
Cited by
-
Comparative analysis of Cd-responsive maize and rice transcriptomes highlights Cd co-modulated orthologs.BMC Genomics. 2018 Sep 26;19(1):709. doi: 10.1186/s12864-018-5109-8. BMC Genomics. 2018. PMID: 30257650 Free PMC article.
-
BrpNAC895 and BrpABI449 coregulate the transcription of the afflux-type Cd transporter BrpHMA2 in Brassica parachinensis.Hortic Res. 2022 Feb 19;9:uhac044. doi: 10.1093/hr/uhac044. Online ahead of print. Hortic Res. 2022. PMID: 35184182 Free PMC article.
-
Transcriptomics and metabolomics association analysis revealed the responses of Gynostemma pentaphyllum to cadmium.Front Plant Sci. 2023 Oct 9;14:1265971. doi: 10.3389/fpls.2023.1265971. eCollection 2023. Front Plant Sci. 2023. PMID: 37877087 Free PMC article.
-
An AC-Rich Bean Element Serves as an Ethylene-Responsive Element in Arabidopsis.Plants (Basel). 2020 Aug 14;9(8):1033. doi: 10.3390/plants9081033. Plants (Basel). 2020. PMID: 32823972 Free PMC article.
-
ERF109 of trifoliate orange (Poncirus trifoliata (L.) Raf.) contributes to cold tolerance by directly regulating expression of Prx1 involved in antioxidative process.Plant Biotechnol J. 2019 Jul;17(7):1316-1332. doi: 10.1111/pbi.13056. Epub 2019 Jan 4. Plant Biotechnol J. 2019. PMID: 30575255 Free PMC article.
References
-
- Chai T, Chen Q, Zhang Y, Dong J, An C (2003) Cadmium resistance in transgenic tobacco plants enhanced by expressing bean heavy metal-responsive gene PvSR2. Sci China C Life Sci 46: 623–630 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

