Major Shifts in Glial Regional Identity Are a Transcriptional Hallmark of Human Brain Aging
- PMID: 28076797
- PMCID: PMC5263238
- DOI: 10.1016/j.celrep.2016.12.011
Major Shifts in Glial Regional Identity Are a Transcriptional Hallmark of Human Brain Aging
Abstract
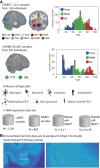
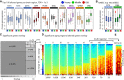
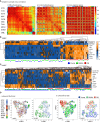
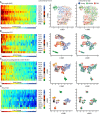
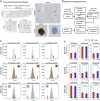
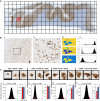
Gene expression studies suggest that aging of the human brain is determined by a complex interplay of molecular events, although both its region- and cell-type-specific consequences remain poorly understood. Here, we extensively characterized aging-altered gene expression changes across ten human brain regions from 480 individuals ranging in age from 16 to 106 years. We show that astrocyte- and oligodendrocyte-specific genes, but not neuron-specific genes, shift their regional expression patterns upon aging, particularly in the hippocampus and substantia nigra, while the expression of microglia- and endothelial-specific genes increase in all brain regions. In line with these changes, high-resolution immunohistochemistry demonstrated decreased numbers of oligodendrocytes and of neuronal subpopulations in the aging brain cortex. Finally, glial-specific genes predict age with greater precision than neuron-specific genes, thus highlighting the need for greater mechanistic understanding of neuron-glia interactions in aging and late-life diseases.
Keywords: RNA-seq; aging; brain; exon microarrays; gene expression; immunohistochemistry; machine learning; microglia; neurons; olgiodendrocytes.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

