SCOPA and META-SCOPA: software for the analysis and aggregation of genome-wide association studies of multiple correlated phenotypes
- PMID: 28077070
- PMCID: PMC5225593
- DOI: 10.1186/s12859-016-1437-3
SCOPA and META-SCOPA: software for the analysis and aggregation of genome-wide association studies of multiple correlated phenotypes
Abstract
Background: Genome-wide association studies (GWAS) of single nucleotide polymorphisms (SNPs) have been successful in identifying loci contributing genetic effects to a wide range of complex human diseases and quantitative traits. The traditional approach to GWAS analysis is to consider each phenotype separately, despite the fact that many diseases and quantitative traits are correlated with each other, and often measured in the same sample of individuals. Multivariate analyses of correlated phenotypes have been demonstrated, by simulation, to increase power to detect association with SNPs, and thus may enable improved detection of novel loci contributing to diseases and quantitative traits.
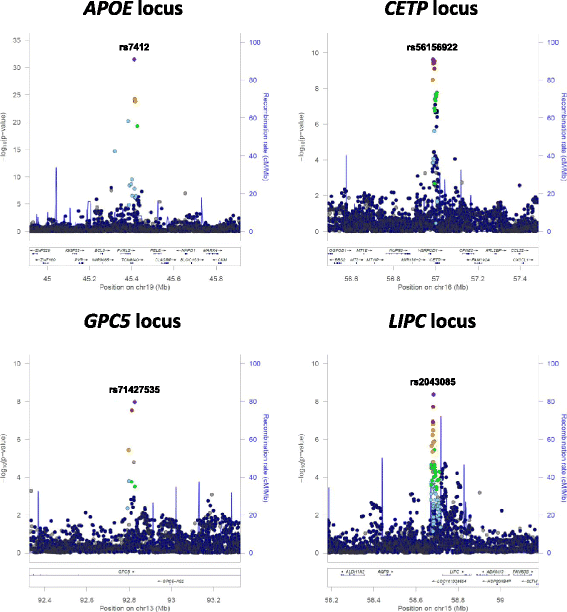
Results: We have developed the SCOPA software to enable GWAS analysis of multiple correlated phenotypes. The software implements "reverse regression" methodology, which treats the genotype of an individual at a SNP as the outcome and the phenotypes as predictors in a general linear model. SCOPA can be applied to quantitative traits and categorical phenotypes, and can accommodate imputed genotypes under a dosage model. The accompanying META-SCOPA software enables meta-analysis of association summary statistics from SCOPA across GWAS. Application of SCOPA to two GWAS of high-and low-density lipoprotein cholesterol, triglycerides and body mass index, and subsequent meta-analysis with META-SCOPA, highlighted stronger association signals than univariate phenotype analysis at established lipid and obesity loci. The META-SCOPA meta-analysis also revealed a novel signal of association at genome-wide significance for triglycerides mapping to GPC5 (lead SNP rs71427535, p = 1.1x10-8), which has not been reported in previous large-scale GWAS of lipid traits.
Conclusions: The SCOPA and META-SCOPA software enable discovery and dissection of multiple phenotype association signals through implementation of a powerful reverse regression approach.
Keywords: Correlation; Genome-wide association study; Meta-analysis; Multiple phenotypes; Multivariate analysis; Reverse regression.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

