Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain
- PMID: 28077169
- PMCID: PMC5225599
- DOI: 10.1186/s13059-016-1139-1
Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain
Abstract
Background: Recent work has identified and mapped a range of posttranscriptional modifications in mRNA, including methylation of the N6 and N1 positions in adenine, pseudouridylation, and methylation of carbon 5 in cytosine (m5C). However, knowledge about the prevalence and transcriptome-wide distribution of m5C is still extremely limited; thus, studies in different cell types, tissues, and organisms are needed to gain insight into possible functions of this modification and implications for other regulatory processes.
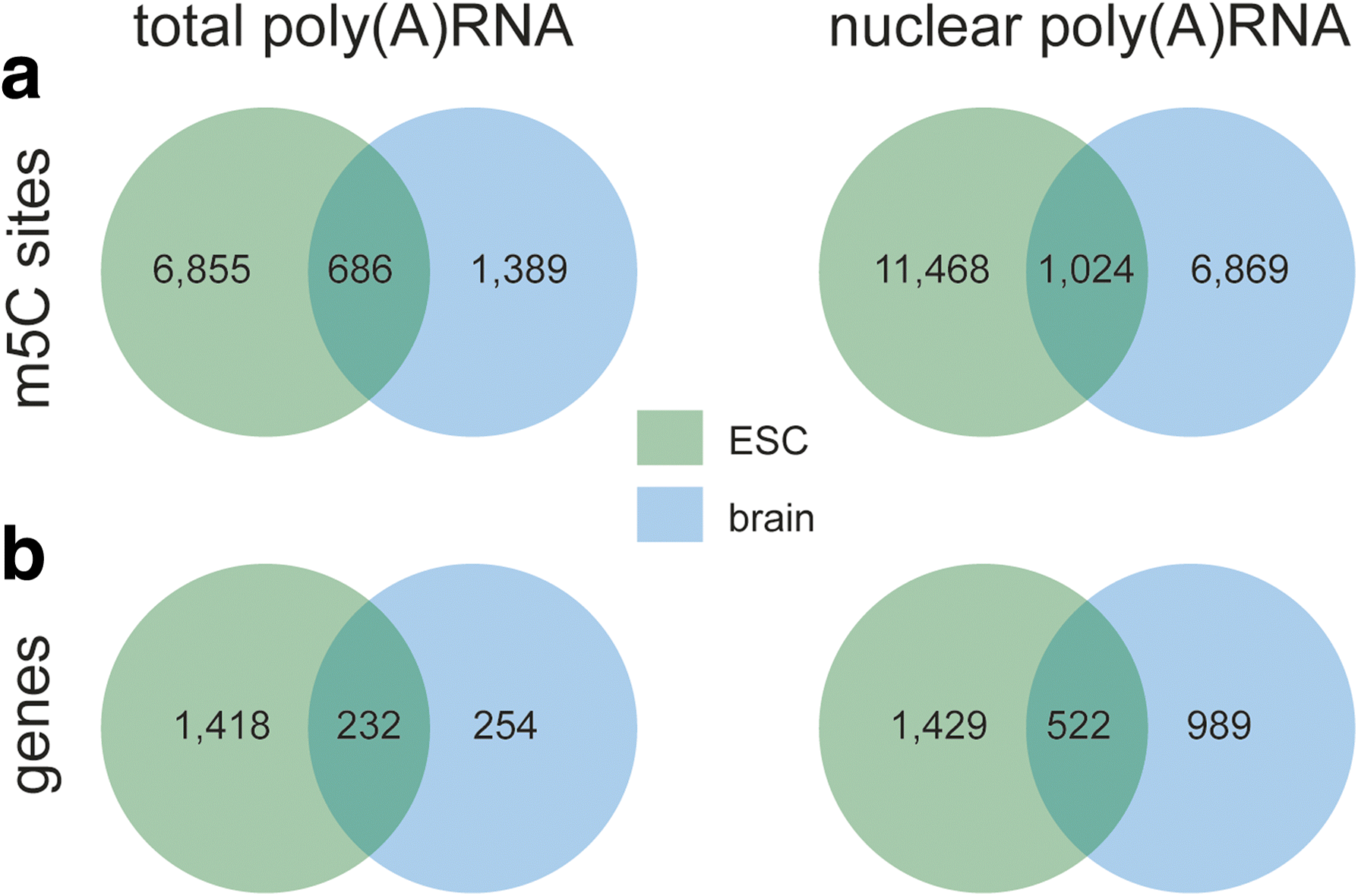
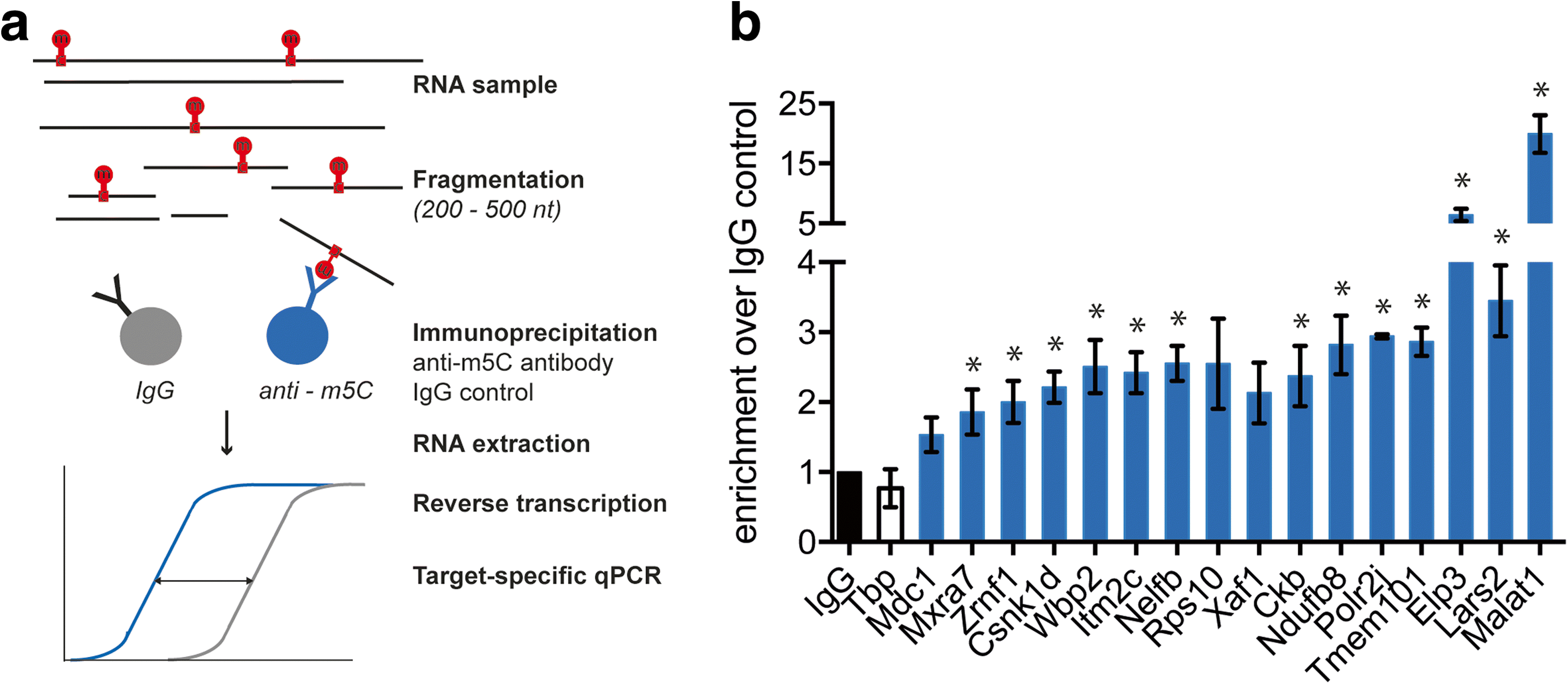
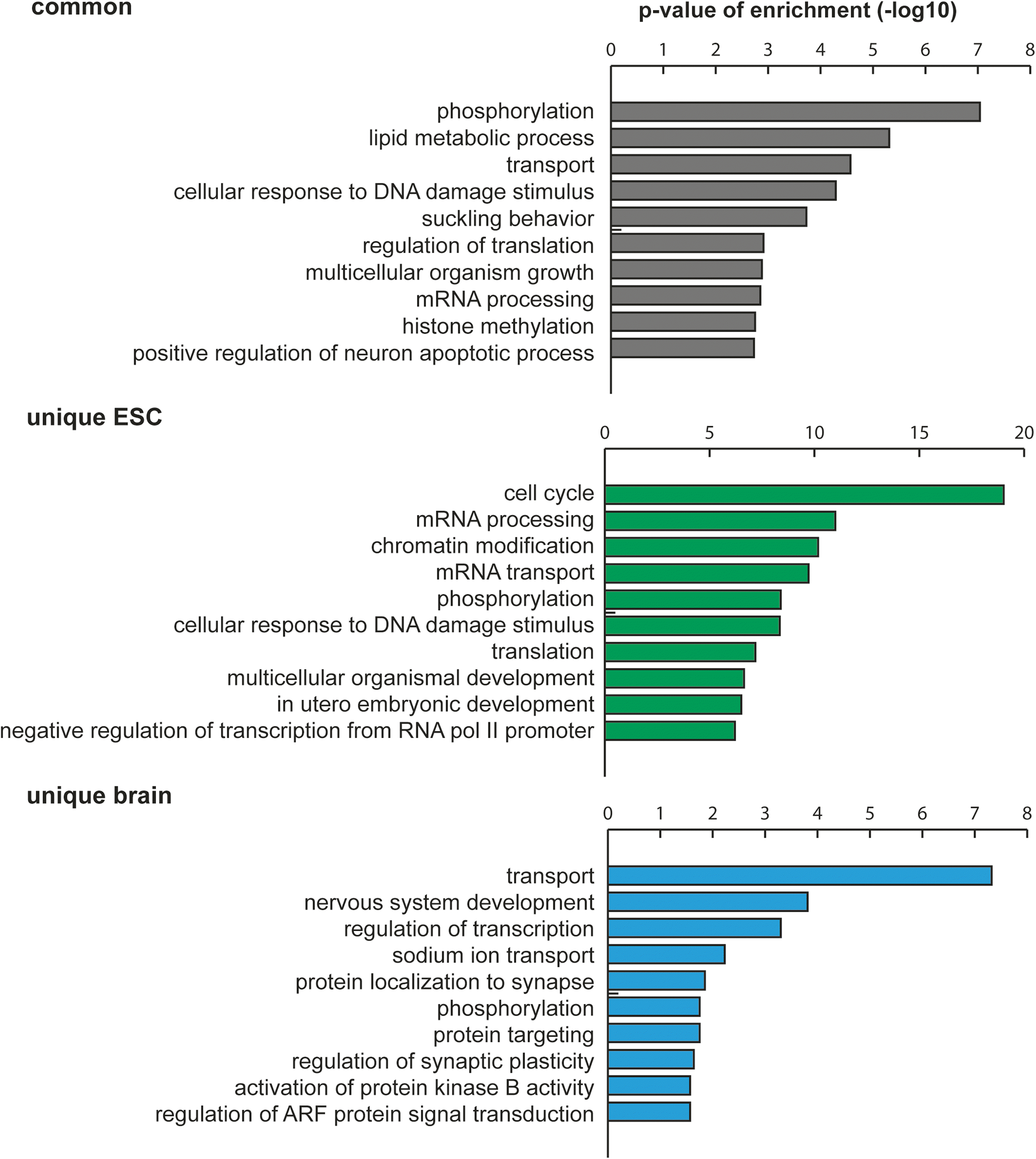
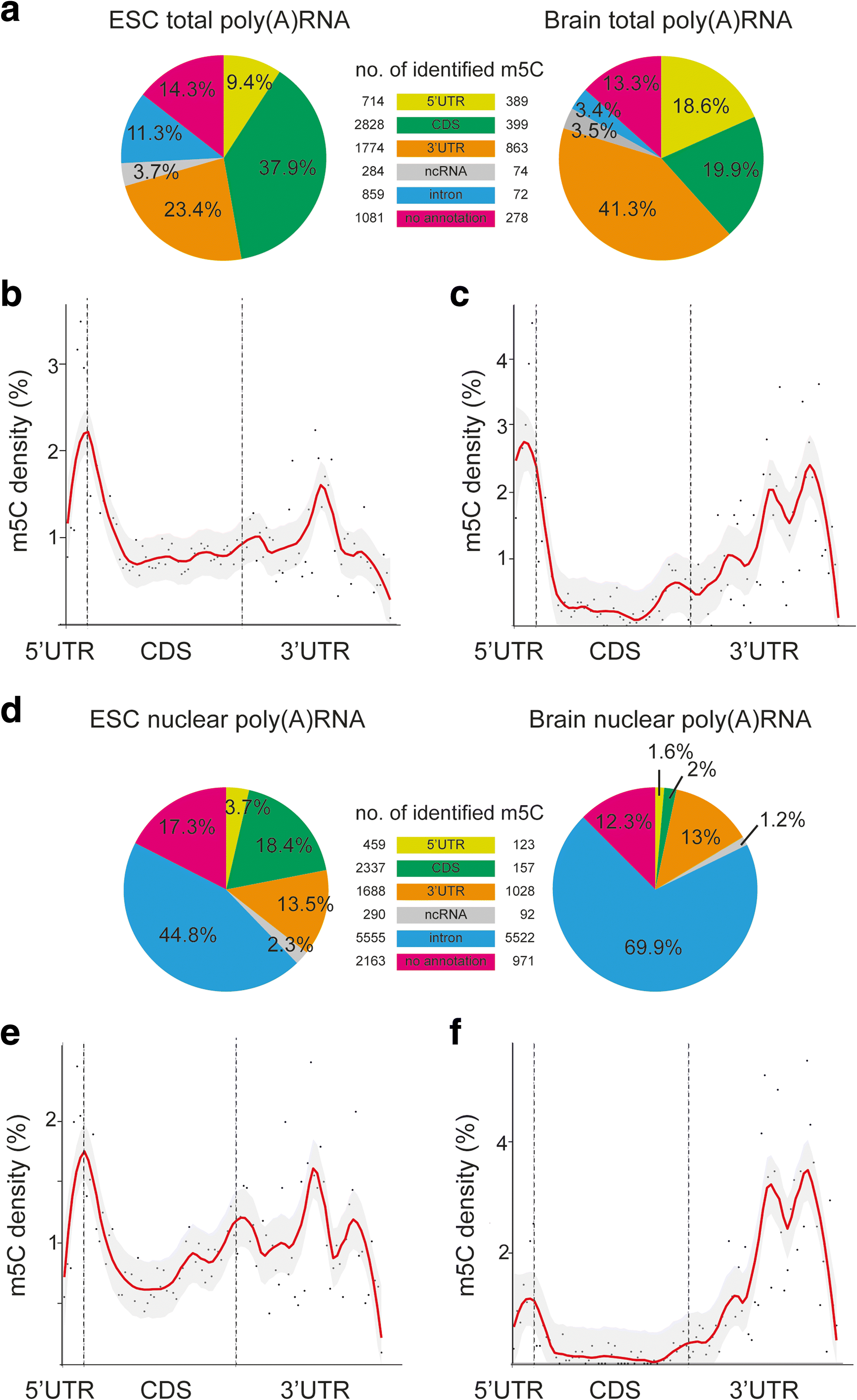
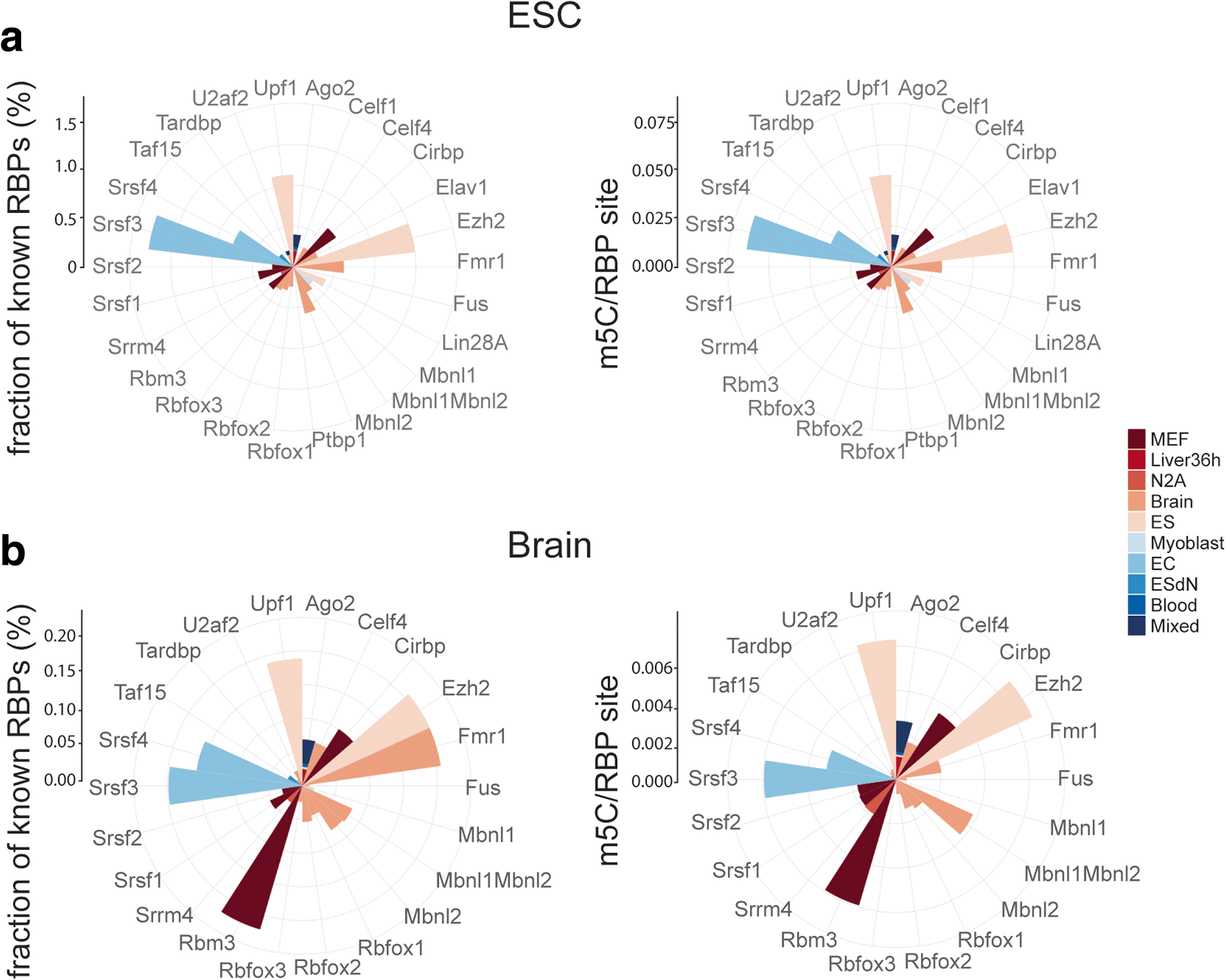
Results: We have carried out an unbiased global analysis of m5C in total and nuclear poly(A) RNA of mouse embryonic stem cells and murine brain. We show that there are intriguing differences in these samples and cell compartments with respect to the degree of methylation, functional classification of methylated transcripts, and position bias within the transcript. Specifically, we observe a pronounced accumulation of m5C sites in the vicinity of the translational start codon, depletion in coding sequences, and mixed patterns of enrichment in the 3' UTR. Degree and pattern of methylation distinguish transcripts modified in both embryonic stem cells and brain from those methylated in either one of the samples. We also analyze potential correlations between m5C and micro RNA target sites, binding sites of RNA binding proteins, and N6-methyladenosine.
Conclusion: Our study presents the first comprehensive picture of cytosine methylation in the epitranscriptome of pluripotent and differentiated stages in the mouse. These data provide an invaluable resource for future studies of function and biological significance of m5C in mRNA in mammals.
Keywords: 5-Methylcytosine; Bisulfite sequencing; Embryonic stem cells; Epitranscriptome; Mouse brain; RNA binding proteins; RNA methylation; m5C; m6A; meRIP.
Figures
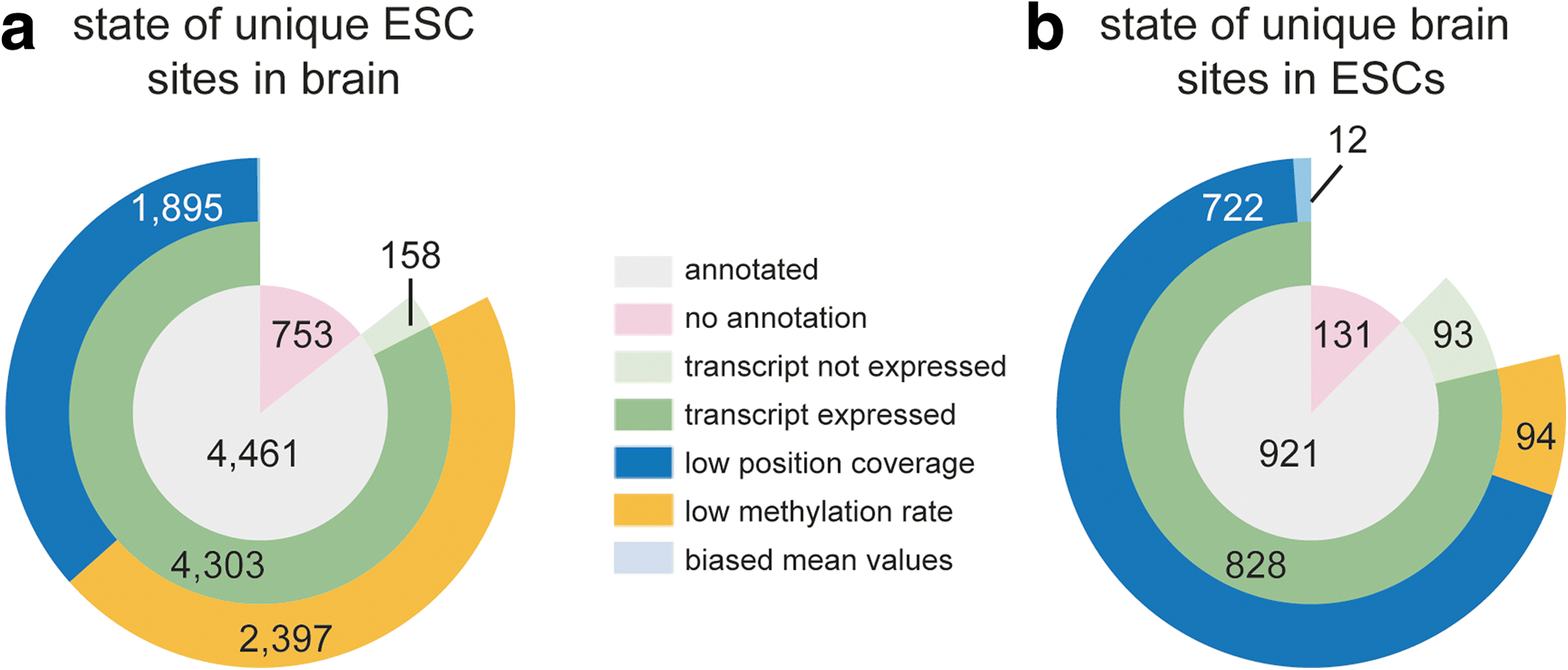
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

