Human Antiviral Protein IFIX Suppresses Viral Gene Expression during Herpes Simplex Virus 1 (HSV-1) Infection and Is Counteracted by Virus-induced Proteasomal Degradation
- PMID: 28077445
- PMCID: PMC5393386
- DOI: 10.1074/mcp.M116.064741
Human Antiviral Protein IFIX Suppresses Viral Gene Expression during Herpes Simplex Virus 1 (HSV-1) Infection and Is Counteracted by Virus-induced Proteasomal Degradation
Abstract
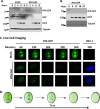
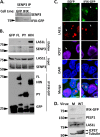
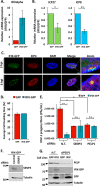
The interferon-inducible protein X (IFIX), a member of the PYHIN family, was recently recognized as an antiviral factor against infection with herpes simplex virus 1 (HSV-1). IFIX binds viral DNA upon infection and promotes expression of antiviral cytokines. How IFIX exerts its host defense functions and whether it is inhibited by the virus remain unknown. Here, we integrated live cell microscopy, proteomics, IFIX domain characterization, and molecular virology to investigate IFIX regulation and antiviral functions during HSV-1 infection. We find that IFIX has a dynamic localization during infection that changes from diffuse nuclear and nucleoli distribution in uninfected cells to discrete nuclear puncta early in infection. This is rapidly followed by a reduction in IFIX protein levels. Indeed, using immunoaffinity purification and mass spectrometry, we define IFIX interactions during HSV-1 infection, finding an association with a proteasome subunit and proteins involved in ubiquitin-proteasome processes. Using synchronized HSV-1 infection, microscopy, and proteasome-inhibition experiments, we demonstrate that IFIX co-localizes with nuclear proteasome puncta shortly after 3 h of infection and that its pyrin domain is rapidly degraded in a proteasome-dependent manner. We further demonstrate that, in contrast to several other host defense factors, IFIX degradation is not dependent on the E3 ubiquitin ligase activity of the viral protein ICP0. However, we show IFIX degradation requires immediate-early viral gene expression, suggesting a viral host suppression mechanism. The IFIX interactome also demonstrated its association with transcriptional regulatory proteins, including the 5FMC complex. We validate this interaction using microscopy and reciprocal isolations and determine it is mediated by the IFIX HIN domain. Finally, we show IFIX suppresses immediate-early and early viral gene expression during infection. Altogether, our study demonstrates that IFIX antiviral functions work in part via viral transcriptional suppression and that HSV-1 has acquired mechanisms to block its functions via proteasome-dependent degradation.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

