Regulating chromosomal movement by the cochaperone FKB-6 ensures timely pairing and synapsis
- PMID: 28077446
- PMCID: PMC5294783
- DOI: 10.1083/jcb.201606126
Regulating chromosomal movement by the cochaperone FKB-6 ensures timely pairing and synapsis
Abstract
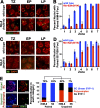
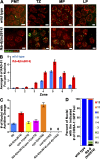
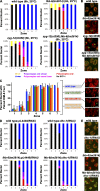
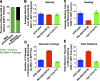
In meiotic prophase I, homologous chromosome pairing is promoted through chromosome movement mediated by nuclear envelope proteins, microtubules, and dynein. After proper homologue pairing has been established, the synaptonemal complex (SC) assembles along the paired homologues, stabilizing their interaction and allowing for crossing over to occur. Previous studies have shown that perturbing chromosome movement leads to pairing defects and SC polycomplex formation. We show that FKB-6 plays a role in SC assembly and is required for timely pairing and proper double-strand break repair kinetics. FKB-6 localizes outside the nucleus, and in its absence, the microtubule network is altered. FKB-6 is required for proper movement of dynein, increasing resting time between movements. Attenuating chromosomal movement in fkb-6 mutants partially restores the defects in synapsis, in agreement with FKB-6 acting by decreasing chromosomal movement. Therefore, we suggest that FKB-6 plays a role in regulating dynein movement by preventing excess chromosome movement, which is essential for proper SC assembly and homologous chromosome pairing.
© 2017 Alleva et al.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

