RAIN: RNA-protein Association and Interaction Networks
- PMID: 28077569
- PMCID: PMC5225963
- DOI: 10.1093/database/baw167
RAIN: RNA-protein Association and Interaction Networks
Erratum in
-
Correction notice for booking in.Database (Oxford). 2017 Jan 1;2017(1):bax007. doi: 10.1093/database/bax007. Database (Oxford). 2017. PMID: 28365717 Free PMC article. No abstract available.
Abstract
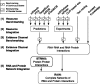
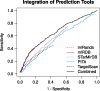
Protein association networks can be inferred from a range of resources including experimental data, literature mining and computational predictions. These types of evidence are emerging for non-coding RNAs (ncRNAs) as well. However, integration of ncRNAs into protein association networks is challenging due to data heterogeneity. Here, we present a database of ncRNA-RNA and ncRNA-protein interactions and its integration with the STRING database of protein-protein interactions. These ncRNA associations cover four organisms and have been established from curated examples, experimental data, interaction predictions and automatic literature mining. RAIN uses an integrative scoring scheme to assign a confidence score to each interaction. We demonstrate that RAIN outperforms the underlying microRNA-target predictions in inferring ncRNA interactions. RAIN can be operated through an easily accessible web interface and all interaction data can be downloaded.Database URL: http://rth.dk/resources/rain.
© The Author(s) 2017. Published by Oxford University Press.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

