Neuron-enriched RNA-binding Proteins Regulate Pancreatic Beta Cell Function and Survival
- PMID: 28077579
- PMCID: PMC5336178
- DOI: 10.1074/jbc.M116.748335
Neuron-enriched RNA-binding Proteins Regulate Pancreatic Beta Cell Function and Survival
Abstract
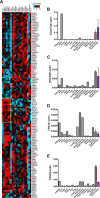
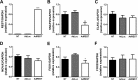
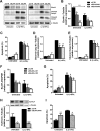
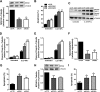
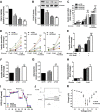
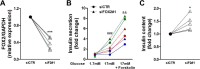
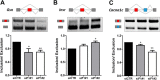
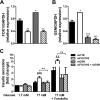
Pancreatic beta cell failure is the central event leading to diabetes. Beta cells share many phenotypic traits with neurons, and proper beta cell function relies on the activation of several neuron-like transcription programs. Regulation of gene expression by alternative splicing plays a pivotal role in brain, where it affects neuronal development, function, and disease. The role of alternative splicing in beta cells remains unclear, but recent data indicate that splicing alterations modulated by both inflammation and susceptibility genes for diabetes contribute to beta cell dysfunction and death. Here we used RNA sequencing to compare the expression of splicing-regulatory RNA-binding proteins in human islets, brain, and other human tissues, and we identified a cluster of splicing regulators that are expressed in both beta cells and brain. Four of them, namely Elavl4, Nova2, Rbox1, and Rbfox2, were selected for subsequent functional studies in insulin-producing rat INS-1E, human EndoC-βH1 cells, and in primary rat beta cells. Silencing of Elavl4 and Nova2 increased beta cell apoptosis, whereas silencing of Rbfox1 and Rbfox2 increased insulin content and secretion. Interestingly, Rbfox1 silencing modulates the splicing of the actin-remodeling protein gelsolin, increasing gelsolin expression and leading to faster glucose-induced actin depolymerization and increased insulin release. Taken together, these findings indicate that beta cells share common splicing regulators and programs with neurons. These splicing regulators play key roles in insulin release and beta cell survival, and their dysfunction may contribute to the loss of functional beta cell mass in diabetes.
Keywords: alternative splicing; apoptosis; autoimmunity; beta cell (B-cell); diabetes; insulin secretion.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

References
-
- Arntfield M. E., and van der Kooy D. (2011) Beta-cell evolution: how the pancreas borrowed from the brain. The shared toolbox of genes expressed by neural and pancreatic endocrine cells may reflect their evolutionary relationship. Bioessays 33, 582–587 - PubMed
-
- Regazzi R., Wollheim C. B., Lang J., Theler J. M., Rossetto O., Montecucco C., Sadoul K., Weller U., Palmer M., and Thorens B. (1995) VAMP-2 and cellubrevin are expressed in pancreatic beta-cells and are essential for Ca2+-but not for GTPγS-induced insulin secretion. EMBO J. 14, 2723–2730 - PMC - PubMed
-
- Leung Y. M., Kwan E. P., Ng B., Kang Y., and Gaisano H. Y. (2007) SNAREing voltage-gated K+ and ATP-sensitive K+ channels: tuning beta-cell excitability with syntaxin-1A and other exocytotic proteins. Endocr. Rev. 28, 653–663 - PubMed
-
- Henquin J. C., and Meissner H. P. (1984) The ionic, electrical, and secretory effects of endogenous cyclic adenosine monophosphate in mouse pancreatic B cells: studies with forskolin. Endocrinology 115, 1125–1134 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

