Characterizing HIV-1 Splicing by Using Next-Generation Sequencing
- PMID: 28077653
- PMCID: PMC5331825
- DOI: 10.1128/JVI.02515-16
Characterizing HIV-1 Splicing by Using Next-Generation Sequencing
Abstract
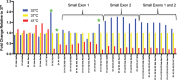
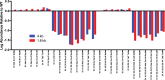
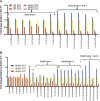
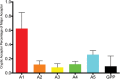
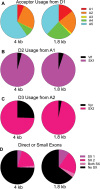
Full-length human immunodeficiency virus type 1 (HIV-1) RNA serves as the genome or as an mRNA, or this RNA undergoes splicing using four donors and 10 acceptors to create over 50 physiologically relevant transcripts in two size classes (1.8 kb and 4 kb). We developed an assay using Primer ID-tagged deep sequencing to quantify HIV-1 splicing. Using the lab strain NL4-3, we found that A5 (env/nef) is the most commonly used acceptor (about 50%) and A3 (tat) the least used (about 3%). Two small exons are made when a splice to acceptor A1 or A2 is followed by activation of donor D2 or D3, and the high-level use of D2 and D3 dramatically reduces the amount of vif and vpr transcripts. We observed distinct patterns of temperature sensitivity of splicing to acceptors A1 and A2. In addition, disruption of a conserved structure proximal to A1 caused a 10-fold reduction in all transcripts that utilized A1. Analysis of a panel of subtype B transmitted/founder viruses showed that splicing patterns are conserved, but with surprising variability of usage. A subtype C isolate was similar, while a simian immunodeficiency virus (SIV) isolate showed significant differences. We also observed transsplicing from a downstream donor on one transcript to an upstream acceptor on a different transcript, which we detected in 0.3% of 1.8-kb RNA reads. There were several examples of splicing suppression when the env intron was retained in the 4-kb size class. These results demonstrate the utility of this assay and identify new examples of HIV-1 splicing regulation. IMPORTANCE During HIV-1 replication, over 50 conserved spliced RNA variants are generated. The splicing assay described here uses new developments in deep-sequencing technology combined with Primer ID-tagged cDNA primers to efficiently quantify HIV-1 splicing at a depth that allows even low-frequency splice variants to be monitored. We have used this assay to examine several features of HIV-1 splicing and to identify new examples of different mechanisms of regulation of these splicing patterns. This splicing assay can be used to explore in detail how HIV-1 splicing is regulated and, with moderate throughput, could be used to screen for structural elements, small molecules, and host factors that alter these relatively conserved splicing patterns.
Keywords: HIV-1; RNA splicing; next-generation sequencing; primer ID; simian immunodeficiency virus.
Copyright © 2017 American Society for Microbiology.
Figures



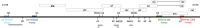

References
-
- Ocwieja KE, Sherrill-Mix S, Mukherjee R, Custers-Allen R, David P, Brown M, Wang S, Link DR, Olson J, Travers K, Schadt E, Bushman FD. 2012. Dynamic regulation of HIV-1 mRNA populations analyzed by single-molecule enrichment and long-read sequencing. Nucleic Acids Res 40:10345–10355. doi: 10.1093/nar/gks753. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

