Integration of temporal and spatial patterning generates neural diversity
- PMID: 28077877
- PMCID: PMC5489111
- DOI: 10.1038/nature20794
Integration of temporal and spatial patterning generates neural diversity
Abstract
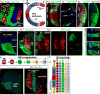
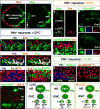
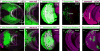
In the Drosophila optic lobes, 800 retinotopically organized columns in the medulla act as functional units for processing visual information. The medulla contains over 80 types of neuron, which belong to two classes: uni-columnar neurons have a stoichiometry of one per column, while multi-columnar neurons contact multiple columns. Here we show that combinatorial inputs from temporal and spatial axes generate this neuronal diversity: all neuroblasts switch fates over time to produce different neurons; the neuroepithelium that generates neuroblasts is also subdivided into six compartments by the expression of specific factors. Uni-columnar neurons are produced in all spatial compartments independently of spatial input; they innervate the neuropil where they are generated. Multi-columnar neurons are generated in smaller numbers in restricted compartments and require spatial input; the majority of their cell bodies subsequently move to cover the entire medulla. The selective integration of spatial inputs by a fixed temporal neuroblast cascade thus acts as a powerful mechanism for generating neural diversity, regulating stoichiometry and the formation of retinotopy.
Figures



References
-
- Fischbach KF, D APM. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 1989;258:441–475.
-
- Bausenwein B, Dittrich AP, Fischbach KF. The optic lobe of Drosophila melanogaster. II. Sorting of retinotopic pathways in the medulla. Cell Tissue Res. 1992;267:17–28. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

