Artificial cells: from basic science to applications
- PMID: 28077925
- PMCID: PMC5222523
- DOI: 10.1016/j.mattod.2016.02.020
Artificial cells: from basic science to applications
Abstract
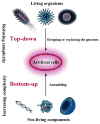
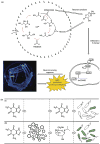
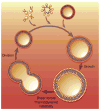
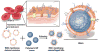
Artificial cells have attracted much attention as substitutes for natural cells. There are many different forms of artificial cells with many different definitions. They can be integral biological cell imitators with cell-like structures and exhibit some of the key characteristics of living cells. Alternatively, they can be engineered materials that only mimic some of the properties of cells, such as surface characteristics, shapes, morphology, or a few specific functions. These artificial cells can have applications in many fields from medicine to environment, and may be useful in constructing the theory of the origin of life. However, even the simplest unicellular organisms are extremely complex and synthesis of living artificial cells from inanimate components seems very daunting. Nevertheless, recent progress in the formulation of artificial cells ranging from simple protocells and synthetic cells to cell-mimic particles, suggests that the construction of living life is now not an unrealistic goal. This review aims to provide a comprehensive summary of the latest developments in the construction and application of artificial cells, as well as highlight the current problems, limitations, challenges and opportunities in this field.
Figures








References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
