An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites
- PMID: 28079019
- PMCID: PMC5310842
- DOI: 10.7554/eLife.21856
An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites
Abstract
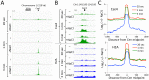
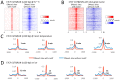
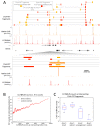
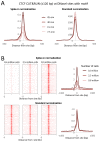
We describe Cleavage Under Targets and Release Using Nuclease (CUT&RUN), a chromatin profiling strategy in which antibody-targeted controlled cleavage by micrococcal nuclease releases specific protein-DNA complexes into the supernatant for paired-end DNA sequencing. Unlike Chromatin Immunoprecipitation (ChIP), which fragments and solubilizes total chromatin, CUT&RUN is performed in situ, allowing for both quantitative high-resolution chromatin mapping and probing of the local chromatin environment. When applied to yeast and human nuclei, CUT&RUN yielded precise transcription factor profiles while avoiding crosslinking and solubilization issues. CUT&RUN is simple to perform and is inherently robust, with extremely low backgrounds requiring only ~1/10th the sequencing depth as ChIP, making CUT&RUN especially cost-effective for transcription factor and chromatin profiling. When used in conjunction with native ChIP-seq and applied to human CTCF, CUT&RUN mapped directional long range contact sites at high resolution. We conclude that in situ mapping of protein-DNA interactions by CUT&RUN is an attractive alternative to ChIP-seq.
Keywords: DNA sequencing; S. cerevisiae; chromatin; chromosomes; genes; human; in situ profiling; transcription factors.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures

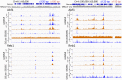
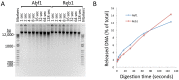

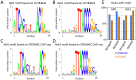
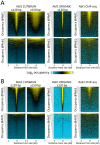













Comment in
-
A cut above.Elife. 2017 Feb 15;6:e25000. doi: 10.7554/eLife.25000. Elife. 2017. PMID: 28199181 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

