Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1
- PMID: 28080204
- PMCID: PMC5367248
- DOI: 10.1080/15476286.2017.1279788
Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1
Abstract
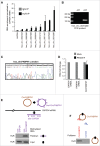
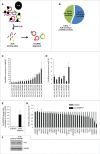
HuR influences gene expression programs and hence cellular phenotypes by binding to hundreds of coding and noncoding linear RNAs. However, whether HuR binds to circular RNAs (circRNAs) and impacts on their function is unknown. Here, we have identified en masse circRNAs binding HuR in human cervical carcinoma HeLa cells. One of the most prominent HuR target circRNAs was hsa_circ_0031288, renamed CircPABPN1 as it arises from the PABPN1 pre-mRNA. Further analysis revealed that HuR did not influence CircPABPN1 abundance; interestingly, however, high levels of CircPABPN1 suppressed HuR binding to PABPN1 mRNA. Evaluation of PABPN1 mRNA polysomes indicated that PABPN1 translation was modulated positively by HuR and hence negatively by CircPABPN1. We propose that the extensive binding of CircPABPN1 to HuR prevents HuR binding to PABPN1 mRNA and lowers PABPN1 translation, providing the first example of competition between a circRNA and its cognate mRNA for an RBP that affects translation.
Keywords: Cell proliferation; HuR; PABPN1; RNA-binding Protein; circPABPN1; circular RNA; endogenous competing RNA; translation.
Figures


References
-
- Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell 2011; 43:340-52; PMID:21723171; http://dx.doi.org/ 10.1016/j.molcel.2011.06.008 - DOI - PubMed
-
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell 2011; 43:327-39; PMID:21723170; http://dx.doi.org/ 10.1016/j.molcel.2011.06.007 - DOI - PMC - PubMed
-
- Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem 2008; 389:243-55; PMID:18177264; http://dx.doi.org/ 10.1515/BC.2008.022 - DOI - PMC - PubMed
-
- Srikantan S, Gorospe M. HuR function in disease. Front Biosci (Landmark Ed) 2012; 17:189-205; PMID:22201738; http://dx.doi.org/ 10.2741/3921 - DOI - PMC - PubMed
-
- Grammatikakis I, Abdelmohsen K, Gorospe M. Posttranslational control of HuR function.Wiley Interdiscip Rev RNA; 2017; 8(1); PMID: 27307117; http://dx.doi.org/23178169 10.1002/wrna.1372 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
