Human Lacrimal Gland Gene Expression
- PMID: 28081151
- PMCID: PMC5231359
- DOI: 10.1371/journal.pone.0169346
Human Lacrimal Gland Gene Expression
Abstract
Background: The study of human lacrimal gland biology and development is limited. Lacrimal gland tissue is damaged or poorly functional in a number of disease states including dry eye disease. Development of cell based therapies for lacrimal gland diseases requires a better understanding of the gene expression and signaling pathways in lacrimal gland. Differential gene expression analysis between lacrimal gland and other embryologically similar tissues may be helpful in furthering our understanding of lacrimal gland development.
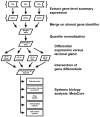
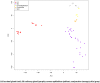
Methods: We performed global gene expression analysis of human lacrimal gland tissue using Affymetrix ® gene expression arrays. Primary data from our laboratory was compared with datasets available in the NLM GEO database for other surface ectodermal tissues including salivary gland, skin, conjunctiva and corneal epithelium.
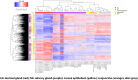
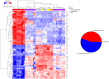
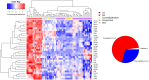
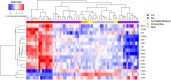
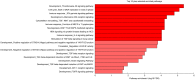
Results: The analysis revealed statistically significant difference in the gene expression of lacrimal gland tissue compared to other ectodermal tissues. The lacrimal gland specific, cell surface secretory protein encoding genes and critical signaling pathways which distinguish lacrimal gland from other ectodermal tissues are described.
Conclusions: Differential gene expression in human lacrimal gland compared with other ectodermal tissue types revealed interesting patterns which may serve as the basis for future studies in directed differentiation among other areas.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- The epidemiology of dry eye disease: report of the Epidemiology Subcommittee of the International Dry Eye WorkShop. Ocul Surf. 2007;5(2):93–107. - PubMed
-
- Voronov D, Gromova A, Liu D, Zoukhri D, Medvinsky A, Meech R, et al. Transcription factors Runx1 to 3 are expressed in the lacrimal gland epithelium and are involved in regulation of gland morphogenesis and regeneration. Invest Ophthalmol Vis Sci. 2013;54(5):3115–25. 10.1167/iovs.13-11791 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

