Bacterial Communities Associated with Houseflies (Musca domestica L.) Sampled within and between Farms
- PMID: 28081167
- PMCID: PMC5232358
- DOI: 10.1371/journal.pone.0169753
Bacterial Communities Associated with Houseflies (Musca domestica L.) Sampled within and between Farms
Abstract
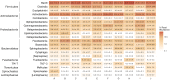
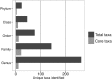
The housefly feeds and reproduces in animal manure and decaying organic substances and thus lives in intimate association with various microorganisms including human pathogens. In order to understand the variation and association between bacteria and the housefly, we used 16S rRNA gene amplicon sequencing to describe bacterial communities of 90 individual houseflies collected within and between ten dairy farms in Denmark. Analysis of gene sequences showed that the most abundant classes of bacteria found across all sites included Bacilli, Clostridia, Actinobacteria, Flavobacteria, and all classes of Proteobacteria and at the genus level the most abundant genera included Corynebacterium, Lactobacillus, Staphylococcus, Vagococcus, Weissella, Lactococcus, and Aerococcus. Comparison of the microbiota of houseflies revealed a highly diverse microbiota compared to other insect species and with most variation in species richness and diversity found between individuals, but not locations. Our study is the first in-depth amplicon sequencing study of the housefly microbiota, and collectively shows that the microbiota of single houseflies is highly diverse and differs between individuals likely to reflect the lifestyle of the housefly. We suggest that these results should be taken into account when addressing the transmission of pathogens by the housefly and assessing the vector competence variation under natural conditions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Bahrndorff S, Gill C, Lowenberger C, Skovgård H, Hald B. The effects of temperature and innate immunity on transmission of Campylobacter jejuni (Campylobacterales: Campylobacteraceae) between life stages of Musca domestica (Diptera: Muscidae). J Med Entomol. 2014;51: 670–677. - PubMed
-
- Bosio CF, Beaty BJ, Black WC IV. Quantitative genetics of vector competence for dengue-2 virus in Aedes aegypti. Am J Trop Med Hyg. 1998;59: 965–970. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

