African Non-Human Primates Host Diverse Enteroviruses
- PMID: 28081564
- PMCID: PMC5233426
- DOI: 10.1371/journal.pone.0169067
African Non-Human Primates Host Diverse Enteroviruses
Abstract
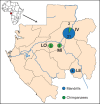
Enteroviruses (EVs) belong to the family Picornaviridae and are responsible for mild to severe diseases in mammals including humans and non-human primates (NHP). Simian EVs were first discovered in the 1950s in the Old World Monkeys and recently in wild chimpanzee, gorilla and mandrill in Cameroon. In the present study, we screened by PCR EVs in 600 fecal samples of wild apes and monkeys that were collected at four sites in Gabon. A total of 32 samples were positive for EVs (25 from mandrills, 7 from chimpanzees, none from gorillas). The phylogenetic analysis of VP1 and VP2 genes showed that EVs identified in chimpanzees were members of two human EV species, EV-A and EV-B, and those identified in mandrills were members of the human species EV-B and the simian species EV-J. The identification of two novel enterovirus types, EV-B112 in a chimpanzee and EV-B113 in a mandrill, suggests these NHPs could be potential sources of new EV types. The identification of EV-B107 and EV90 that were previously found in humans indicates cross-species transfers. Also the identification of chimpanzee-derived EV110 in a mandrill demonstrated a wide host range of this EV. Further research of EVs in NHPs would help understanding emergence of new types or variants, and evaluating the real risk of cross-species transmission for humans as well for NHPs populations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Knowles NJ, Hovi, T, Hyypiä, T., King, A.M.Q., Lindberg, A.M., Pallansch, M.A., Palmenberg, A.C., et al. Family Picornaviridae. In Virus Taxonomy: Classification an Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. 2012:855–80. Ed: King, A.M.Q., Adams, M.J., Carstens, E.B. and Lefkowitz, E.J. San Diego: Elsevier.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

