Defective ATG16L1-mediated removal of IRE1α drives Crohn's disease-like ileitis
- PMID: 28082357
- PMCID: PMC5294857
- DOI: 10.1084/jem.20160791
Defective ATG16L1-mediated removal of IRE1α drives Crohn's disease-like ileitis
Abstract
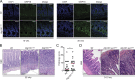
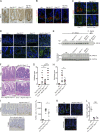
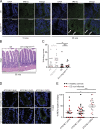
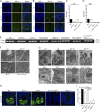
ATG16L1T300A, a major risk polymorphism in Crohn's disease (CD), causes impaired autophagy, but it has remained unclear how this predisposes to CD. In this study, we report that mice with Atg16l1 deletion in intestinal epithelial cells (IECs) spontaneously develop transmural ileitis phenocopying ileal CD in an age-dependent manner, driven by the endoplasmic reticulum (ER) stress sensor IRE1α. IRE1α accumulates in Paneth cells of Atg16l1ΔIEC mice, and humans homozygous for ATG16L1T300A exhibit a corresponding increase of IRE1α in intestinal epithelial crypts. In contrast to a protective role of the IRE1β isoform, hyperactivated IRE1α also drives a similar ileitis developing earlier in life in Atg16l1;Xbp1ΔIEC mice, in which ER stress is induced by deletion of the unfolded protein response transcription factor XBP1. The selective autophagy receptor optineurin interacts with IRE1α, and optineurin deficiency amplifies IRE1α levels during ER stress. Furthermore, although dysbiosis of the ileal microbiota is present in Atg16l1;Xbp1ΔIEC mice as predicted from impaired Paneth cell antimicrobial function, such structural alteration of the microbiota does not trigger ileitis but, rather, aggravates dextran sodium sulfate-induced colitis. Hence, we conclude that defective autophagy in IECs may predispose to CD ileitis via impaired clearance of IRE1α aggregates during ER stress at this site.
© 2017 Tschurtschenthaler et al.
Figures




References
-
- Amthor, H., Otto A., Vulin A., Rochat A., Dumonceaux J., Garcia L., Mouisel E., Hourdé C., Macharia R., Friedrichs M., et al. 2009. Muscle hypertrophy driven by myostatin blockade does not require stem/precursor-cell activity. Proc. Natl. Acad. Sci. USA. 106:7479–7484. 10.1073/pnas.0811129106 - DOI - PMC - PubMed
-
- Anderson, M.J. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26:32–46.
MeSH terms
Substances
Grants and funding
- R37 DK044319/DK/NIDDK NIH HHS/United States
- R01 DK088199/DK/NIDDK NIH HHS/United States
- R01 DK051362/DK/NIDDK NIH HHS/United States
- MR/L000261/1/MRC_/Medical Research Council/United Kingdom
- R01 DK053056/DK/NIDDK NIH HHS/United States
- MR/N001893/1/MRC_/Medical Research Council/United Kingdom
- MR/N001893/1 /MRC_/Medical Research Council/United Kingdom
- P30 DK034854/DK/NIDDK NIH HHS/United States
- 2008/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 200848/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 106260/Z/14/Z /WT_/Wellcome Trust/United Kingdom
- R01 DK044319/DK/NIDDK NIH HHS/United States
- K08 DK093578/DK/NIDDK NIH HHS/United States
- R56 DK053056/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

