Cullin neddylation may allosterically tune polyubiquitin chain length and topology
- PMID: 28082425
- PMCID: PMC7900908
- DOI: 10.1042/BCJ20160748
Cullin neddylation may allosterically tune polyubiquitin chain length and topology
Abstract
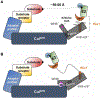
Conjugation of Nedd8 (neddylation) to Cullins (Cul) in Cul-RING E3 ligases (CRLs) stimulates ubiquitination and polyubiquitination of protein substrates. CRL is made up of two Cul-flanked arms: one consists of the substrate-binding and adaptor proteins and the other consists of E2 and Ring-box protein (Rbx). Polyubiquitin chain length and topology determine the substrate fate. Here, we ask how polyubiquitin chains are accommodated in the limited space available between the two arms and what determines the polyubiquitin linkage topology. We focus on Cul5 and Rbx1 in three states: before Cul5 neddylation (closed state), after neddylation (open state), and after deneddylation, exploiting molecular dynamics simulations and the Gaussian Network Model. We observe that regulation of substrate ubiquitination and polyubiquitination takes place through Rbx1 rotations, which are controlled by Nedd8-Rbx1 allosteric communication. Allosteric propagation proceeds from Nedd8 via Cul5 dynamic hinges and hydrogen bonds between the C-terminal domain of Cul5 (Cul5CTD) and Rbx1 (Cul5CTD residues R538/R569 and Rbx1 residue E67, or Cul5CTD E474/E478/N491 and Rbx1 K105). Importantly, at each ubiquitination step (homogeneous or heterogeneous, linear or branched), the polyubiquitin linkages fit into the distances between the two arms, and these match the inherent CRL conformational tendencies. Hinge sites may constitute drug targets.
Keywords: Nedd8; allosteric regulation; polyubiquitination; ubiquitin ligases.
© 2017 The Author(s); published by Portland Press Limited on behalf of the Biochemical Society.
Figures




Similar articles
-
Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation.Cell. 2008 Sep 19;134(6):995-1006. doi: 10.1016/j.cell.2008.07.022. Cell. 2008. PMID: 18805092 Free PMC article.
-
Rbx1 flexible linker facilitates cullin-RING ligase function before neddylation and after deneddylation.Biophys J. 2010 Aug 4;99(3):736-44. doi: 10.1016/j.bpj.2010.05.021. Biophys J. 2010. PMID: 20682250 Free PMC article.
-
Neddylation-induced conformational control regulates cullin RING ligase activity in vivo.J Mol Biol. 2011 Jun 3;409(2):136-45. doi: 10.1016/j.jmb.2011.03.023. Epub 2011 Apr 2. J Mol Biol. 2011. PMID: 21463634
-
Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization.Cell Mol Life Sci. 2009 Jun;66(11-12):1924-38. doi: 10.1007/s00018-009-8712-7. Cell Mol Life Sci. 2009. PMID: 19194658 Free PMC article. Review.
-
Assembly and Regulation of CRL Ubiquitin Ligases.Adv Exp Med Biol. 2020;1217:33-46. doi: 10.1007/978-981-15-1025-0_3. Adv Exp Med Biol. 2020. PMID: 31898220 Review.
Cited by
-
Crystal Structure of the Cul2-Rbx1-EloBC-VHL Ubiquitin Ligase Complex.Structure. 2017 Jun 6;25(6):901-911.e3. doi: 10.1016/j.str.2017.04.009. Structure. 2017. PMID: 28591624 Free PMC article.
-
Characterization of an A3G-VifHIV-1-CRL5-CBFβ Structure Using a Cross-linking Mass Spectrometry Pipeline for Integrative Modeling of Host-Pathogen Complexes.Mol Cell Proteomics. 2021;20:100132. doi: 10.1016/j.mcpro.2021.100132. Epub 2021 Aug 11. Mol Cell Proteomics. 2021. PMID: 34389466 Free PMC article.
-
Development of a BCL-xL and BCL-2 dual degrader with improved anti-leukemic activity.Nat Commun. 2021 Nov 25;12(1):6896. doi: 10.1038/s41467-021-27210-x. Nat Commun. 2021. PMID: 34824248 Free PMC article.
-
Pioneer in Molecular Biology: Conformational Ensembles in Molecular Recognition, Allostery, and Cell Function.J Mol Biol. 2025 Jun 1;437(11):169044. doi: 10.1016/j.jmb.2025.169044. Epub 2025 Feb 25. J Mol Biol. 2025. PMID: 40015370 Review.
-
Comparative Analysis of cul5 and rbx2 Expression in the Developing and Adult Murine Brain and Their Essentiality During Mouse Embryogenesis.Dev Dyn. 2018 Nov;247(11):1227-1236. doi: 10.1002/dvdy.24675. Epub 2018 Nov 7. Dev Dyn. 2018. PMID: 30269386 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

