Pancreas Segmentation in MRI using Graph-Based Decision Fusion on Convolutional Neural Networks
- PMID: 28083570
- PMCID: PMC5223591
- DOI: 10.1007/978-3-319-46723-8_51
Pancreas Segmentation in MRI using Graph-Based Decision Fusion on Convolutional Neural Networks
Abstract
Automated pancreas segmentation in medical images is a prerequisite for many clinical applications, such as diabetes inspection, pancreatic cancer diagnosis, and surgical planing. In this paper, we formulate pancreas segmentation in magnetic resonance imaging (MRI) scans as a graph based decision fusion process combined with deep convolutional neural networks (CNN). Our approach conducts pancreatic detection and boundary segmentation with two types of CNN models respectively: 1) the tissue detection step to differentiate pancreas and non-pancreas tissue with spatial intensity context; 2) the boundary detection step to allocate the semantic boundaries of pancreas. Both detection results of the two networks are fused together as the initialization of a conditional random field (CRF) framework to obtain the final segmentation output. Our approach achieves the mean dice similarity coefficient (DSC) 76.1% with the standard deviation of 8.7% in a dataset containing 78 abdominal MRI scans. The proposed algorithm achieves the best results compared with other state of the arts.
Figures


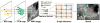

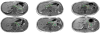
References
-
- Ciresan D, Giusti A, Gambardella LM, Schmidhuber J. Deep neural networks segment neuronal membranes in electron microscopy images. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, editors. Advances in Neural Information Processing Systems. Vol. 25. Curran Associates, Inc; 2012. pp. 2843–2851.
-
- Girshick R, Donahue J, Darrell T, Malik J. Rich feature hierarchies for accurate object detection and semantic segmentation. 2014 IEEE Conference on Computer Vision and Pattern Recognition. 2014 Jun;:580–587.
-
- Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2015 Jun;:3431–3440. - PubMed
-
- Roth HR, Lu L, Farag A, Shin HC, Liu J, Turkbey EB, Summers RM. DeepOrgan: Multi-level Deep Convolutional Networks for Automated Pancreas Segmentation. Springer International Publishing; Cham: 2015. pp. 556–564.
-
- Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. CoRR abs/1409.1556. 2014
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
