Finding Biomass Degrading Enzymes Through an Activity-Correlated Quantitative Proteomics Platform (ACPP)
- PMID: 28083757
- PMCID: PMC5373979
- DOI: 10.1007/s13361-016-1569-8
Finding Biomass Degrading Enzymes Through an Activity-Correlated Quantitative Proteomics Platform (ACPP)
Abstract
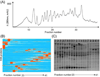
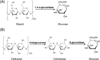
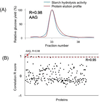
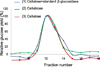
The microbial secretome, known as a pool of biomass (i.e., plant-based materials) degrading enzymes, can be utilized to discover industrial enzyme candidates for biofuel production. Proteomics approaches have been applied to discover novel enzyme candidates through comparing protein expression profiles with enzyme activity of the whole secretome under different growth conditions. However, the activity measurement of each enzyme candidate is needed for confident "active" enzyme assignments, which remains to be elucidated. To address this challenge, we have developed an Activity-Correlated Quantitative Proteomics Platform (ACPP) that systematically correlates protein-level enzymatic activity patterns and protein elution profiles using a label-free quantitative proteomics approach. The ACPP optimized a high performance anion exchange separation for efficiently fractionating complex protein samples while preserving enzymatic activities. The detected enzymatic activity patterns in sequential fractions using microplate-based assays were cross-correlated with protein elution profiles using a customized pattern-matching algorithm with a correlation R-score. The ACPP has been successfully applied to the identification of two types of "active" biomass-degrading enzymes (i.e., starch hydrolysis enzymes and cellulose hydrolysis enzymes) from Aspergillus niger secretome in a multiplexed fashion. By determining protein elution profiles of 156 proteins in A. niger secretome, we confidently identified the 1,4-α-glucosidase as the major "active" starch hydrolysis enzyme (R = 0.96) and the endoglucanase as the major "active" cellulose hydrolysis enzyme (R = 0.97). The results demonstrated that the ACPP facilitated the discovery of bioactive enzymes from complex protein samples in a high-throughput, multiplexing, and untargeted fashion. Graphical Abstract ᅟ.
Keywords: Correlation; Enzyme activity; LC-MS/MS; Peptide counting; Proteomics.
Figures


Similar articles
-
Secretome analysis of Trichoderma reesei and Aspergillus niger cultivated by submerged and sequential fermentation processes: Enzyme production for sugarcane bagasse hydrolysis.Enzyme Microb Technol. 2016 Aug;90:53-60. doi: 10.1016/j.enzmictec.2016.04.011. Epub 2016 Apr 28. Enzyme Microb Technol. 2016. PMID: 27241292
-
Quantitative iTRAQ secretome analysis of Aspergillus niger reveals novel hydrolytic enzymes.J Proteome Res. 2010 Aug 6;9(8):3932-40. doi: 10.1021/pr100148j. J Proteome Res. 2010. PMID: 20545367
-
Heterogeneous Expression and Functional Characterization of Cellulose-Degrading Enzymes from Aspergillus niger for Enzymatic Hydrolysis of Alkali Pretreated Bamboo Biomass.Mol Biotechnol. 2015 Sep;57(9):859-67. doi: 10.1007/s12033-015-9878-x. Mol Biotechnol. 2015. PMID: 26202492
-
Proteomic researches for lignocellulose-degrading enzymes: A mini-review.Bioresour Technol. 2018 Oct;265:532-541. doi: 10.1016/j.biortech.2018.05.101. Epub 2018 May 31. Bioresour Technol. 2018. PMID: 29884341 Review.
-
[Mechanisms and regulation of enzymatic hydrolysis of cellulose in filamentous fungi: classical cases and new models].Rev Iberoam Micol. 2015 Jan-Mar;32(1):1-12. doi: 10.1016/j.riam.2013.10.009. Epub 2014 Mar 7. Rev Iberoam Micol. 2015. PMID: 24607657 Review. Spanish.
Cited by
-
Exploring bioactive peptides from bacterial secretomes using PepSAVI-MS: identification and characterization of Bac-21 from Enterococcus faecalis pPD1.Microb Biotechnol. 2018 Sep;11(5):943-951. doi: 10.1111/1751-7915.13299. Epub 2018 Jul 16. Microb Biotechnol. 2018. PMID: 30014612 Free PMC article.
References
-
- Wu WH, Hung WC, Lo KY, Chen YH, Wan HP, Cheng KC. Bioethanol production from taro waste using thermo-tolerant yeast Kluyveromyces marxianus K21. Bioresour. Technol. 2016;201:27–32. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

