RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway
- PMID: 28085152
- PMCID: PMC5568705
- DOI: 10.1038/nmicrobiol.2016.253
RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway
Erratum in
-
Corrigendum: RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway.Nat Microbiol. 2017 Jan 30;2:17019. doi: 10.1038/nmicrobiol.2017.19. Nat Microbiol. 2017. PMID: 28134912 No abstract available.
Abstract
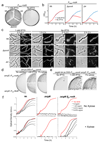
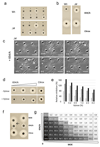
The bacterial cell wall is a highly conserved essential component of most bacterial groups. It is the target for our most frequently used antibiotics and provides important small molecules that trigger powerful innate immune responses. The wall is composed of glycan strands crosslinked by short peptides. For many years, the penicillin-binding proteins were thought to be the key enzymes required for wall synthesis. RodA and possibly other proteins in the wider SEDS (shape, elongation, division and sporulation) family have now emerged as a previously unknown class of essential glycosyltranferase enzymes, which play key morphogenetic roles in bacterial cell wall synthesis. We provide evidence in support of this role and the discovery of small natural product molecules that probably target these enzymes. The SEDS proteins have exceptional potential as targets for new antibacterial therapeutic agents.
Conflict of interest statement
JE is a Director and shareholder at Demuris Ltd. NA and JD are employees at Demuris Ltd.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

