SOX9 chromatin folding domains correlate with its real and putative distant cis-regulatory elements
- PMID: 28085555
- PMCID: PMC5403132
- DOI: 10.1080/19491034.2017.1279776
SOX9 chromatin folding domains correlate with its real and putative distant cis-regulatory elements
Abstract
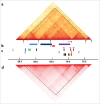
Evolutionary conserved transcription factor SOX9, encoded by the dosage sensitive SOX9 gene on chromosome 17q24.3, plays an important role in development of multiple organs, including bones and testes. Heterozygous point mutations and genomic copy-number variant (CNV) deletions involving SOX9 have been reported in patients with campomelic dysplasia (CD), a skeletal malformation syndrome often associated with male-to-female sex reversal. Balanced and unbalanced structural genomic variants with breakpoints mapping up to 1.3 Mb up- and downstream to SOX9 have been described in patients with milder phenotypes, including acampomelic campomelic dysplasia, sex reversal, and Pierre Robin sequence. Based on the localization of breakpoints of genomic rearrangements causing different phenotypes, 5 genomic intervals mapping upstream to SOX9 have been defined. We have analyzed the publically available database of high-throughput chromosome conformation capture (Hi-C) in multiple cell lines in the genomic regions flanking SOX9. Consistent with the literature data, chromatin domain boundaries in the SOX9 locus exhibit conservation across species and remain largely constant across multiple cell types. Interestingly, we have found that chromatin folding domains in the SOX9 locus associate with the genomic intervals harboring real and putative regulatory elements of SOX9, implicating that variation in intra-domain interactions may be critical for dynamic regulation of SOX9 expression in a cell type-specific fashion. We propose that tissue-specific enhancers for other transcription factor genes may similarly utilize chromatin folding sub-domains in gene regulation.
Keywords: chromatin looping; long distance gene regulation; non-coding variants; structural variants; tissue-specific enhancers.
Figures

References
-
- Gordon CT, Tan TY, Benko S, FitzPatrick D, Lyonnet S, Farlie PG. Long-range regulation at the SOX9 locus in development and disease. J Med Genet 2009; 46:649-56; PMID:19473998; http://dx.doi.org/10.1136/jmg.2009.068361 - DOI - PubMed
-
- Foster JW, Dominguez-Steglich MA, Guioli S, Kwok C, Weller PA, Stevanović M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, et al.. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 1994; 372:525-30; PMID:7990924; http://dx.doi.org/10.1038/372525a0 - DOI - PubMed
-
- Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, et al.. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 1994; 79:1111-20; PMID:8001137; http://dx.doi.org/10.1016/0092-8674(94)90041-8 - DOI - PubMed
-
- Mansour S, Hall CM, Pembrey ME, Young ID. A clinical and genetic study of campomelic dysplasia. J Med Genet 1995; 32:415-20; PMID:7666392; http://jmg.bmj.com.ezproxyhost.library.tmc.edu/content/32/6/415.long - PMC - PubMed
-
- Wirth J, Wagner T, Meyer J, Pfeiffer RA, Tietze HU, Schempp W, Scherer G. Translocation breakpoints in three patients with campomelic dysplasia and autosomal sex reversal map more than 130 kb from SOX9. Hum Genet 1996; 97:186-93; PMID:8566951; http://link.springer.com.ezproxyhost.library.tmc.edu/article/10.1007/BF0... - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
