Immune-Array Analysis in Sporadic Inclusion Body Myositis Reveals HLA-DRB1 Amino Acid Heterogeneity Across the Myositis Spectrum
- PMID: 28086002
- PMCID: PMC5516174
- DOI: 10.1002/art.40045
Immune-Array Analysis in Sporadic Inclusion Body Myositis Reveals HLA-DRB1 Amino Acid Heterogeneity Across the Myositis Spectrum
Abstract
Objective: Inclusion body myositis (IBM) is characterized by a combination of inflammatory and degenerative changes affecting muscle. While the primary cause of IBM is unknown, genetic factors may influence disease susceptibility. To determine genetic factors contributing to the etiology of IBM, we conducted the largest genetic association study of the disease to date, investigating immune-related genes using the Immunochip.
Methods: A total of 252 Caucasian patients with IBM were recruited from 11 countries through the Myositis Genetics Consortium and compared with 1,008 ethnically matched controls. Classic HLA alleles and amino acids were imputed using SNP2HLA.
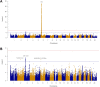
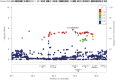
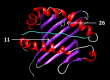
Results: The HLA region was confirmed as the most strongly associated region in IBM (P = 3.58 × 10-33 ). HLA imputation identified 3 independent associations (with HLA-DRB1*03:01, DRB1*01:01, and DRB1*13:01), although the strongest association was with amino acid positions 26 and 11 of the HLA-DRB1 molecule. No association with anti-cytosolic 5'-nucleotidase 1A-positive status was found independent of HLA-DRB1*03:01. There was no association of HLA genotypes with age at onset of IBM. Three non-HLA regions reached suggestive significance, including the chromosome 3 p21.31 region, an established risk locus for autoimmune disease, where a frameshift mutation in CCR5 is thought to be the causal variant.
Conclusion: This is the largest, most comprehensive genetic association study to date in IBM. The data confirm that HLA is the most strongly associated region and identifies novel amino acid associations that may explain the risk in this locus. These amino acid associations differentiate IBM from polymyositis and dermatomyositis and may determine properties of the peptide-binding groove, allowing it to preferentially bind autoantigenic peptides. A novel suggestive association within the chromosome 3 p21.31 region suggests a role for CCR5.
© 2017 The Authors. Arthritis & Rheumatology published by Wiley Periodicals, Inc.on behalf of American College of Rheumatology.
Figures
Comment in
-
Inflammatory myopathies: Genetic associations with IBM.Nat Rev Rheumatol. 2017 Mar;13(3):131. doi: 10.1038/nrrheum.2017.8. Epub 2017 Feb 9. Nat Rev Rheumatol. 2017. PMID: 28202912 No abstract available.
References
-
- Larman HB, Salajegheh M, Nazareno R, Lam T, Sauld J, Steen H, et al. Cytosolic 5′‐nucleotidase 1A autoimmunity in sporadic inclusion body myositis. Ann Neurol 2013;73:408–18. - PubMed
-
- Pluk H, van Hoeve BJ, van Dooren SH, Stammen‐Vogelzangs J, van der Heijden A, Schelhaas HJ, et al. Autoantibodies to cytosolic 5′‐nucleotidase 1A in inclusion body myositis. Ann Neurol 2013;73:397–407. - PubMed
-
- Needham M, Mastaglia FL. Inclusion body myositis: current pathogenetic concepts and diagnostic and therapeutic approaches. Lancet Neurol 2007;6:620–31. - PubMed
-
- Rojana‐udomsart A, James I, Castley A, Needham M, Scott A, Day T, et al. High‐resolution HLA‐DRB1 genotyping in an Australian inclusion body myositis (s‐IBM) cohort: an analysis of disease‐associated alleles and diplotypes. J Neuroimmunol 2012;250:77–82. - PubMed
MeSH terms
Substances
Grants and funding
- MR/K000608/1/MRC_/Medical Research Council/United Kingdom
- 18474/VAC_/Versus Arthritis/United Kingdom
- P30 CA023108/CA/NCI NIH HHS/United States
- MR/K006312/1/MRC_/Medical Research Council/United Kingdom
- MR/K002279/1/MRC_/Medical Research Council/United Kingdom
- MR/N003322/1/MRC_/Medical Research Council/United Kingdom
- 18474/ARC_/Arthritis Research UK/United Kingdom
- MR/P020941/1/MRC_/Medical Research Council/United Kingdom
- G0600237/MRC_/Medical Research Council/United Kingdom
- 105610/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- G0900753/MRC_/Medical Research Council/United Kingdom
- G0100594/MRC_/Medical Research Council/United Kingdom
- G0901461/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

