Identification of regulatory networks and hub genes controlling soybean seed set and size using RNA sequencing analysis
- PMID: 28087653
- PMCID: PMC5429000
- DOI: 10.1093/jxb/erw460
Identification of regulatory networks and hub genes controlling soybean seed set and size using RNA sequencing analysis
Abstract
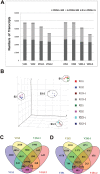
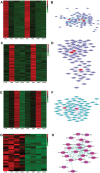
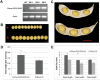
To understand the gene expression networks controlling soybean seed set and size, transcriptome analyses were performed in three early seed developmental stages, using two genotypes with contrasting seed size. The two-dimensional data set provides a comprehensive and systems-level view on dynamic gene expression networks underpinning soybean seed set and subsequent development. Using pairwise comparisons and weighted gene coexpression network analyses, we identified modules of coexpressed genes and hub genes for each module. Of particular importance are the discoveries of specific modules for the large seed size variety and for seed developmental stages. A large number of candidate regulators for seed size, including those involved in hormonal signaling pathways and transcription factors, were transiently and specifically induced in the early developmental stages. The soybean homologs of a brassinosteroid signaling receptor kinase, a brassinosteroid-signaling kinase, were identified as hub genes operating in the seed coat network in the early seed maturation stage. Overexpression of a candidate seed size regulatory gene, GmCYP78A5, in transgenic soybean resulted in increased seed size and seed weight. Together, these analyses identified a large number of potential key regulators controlling soybean seed set, seed size, and, consequently, yield potential, thereby providing new insights into the molecular networks underlying soybean seed development.
Keywords: Seed development; seed number; seed size; soybean; transcriptomic analysis..
© The Author 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures




References
-
- Ainsworth EA, Yendrek CR, Skoneczka JA, Long SP. 2012. Accelerating yield potential in soybean: potential targets for biotechnological improvement. Plant Cell & Environment 35, 38–52. - PubMed
-
- Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, Lenhard M. 2007. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Developmental Cell 13, 843–856. - PubMed
-
- Asakura T, Tamura T, Terauchi K, Narikawa T, Yagasaki K, Ishimaru Y, Abe K. 2012. Global gene expression profiles in developing soybean seeds. Plant Physiology and Biochemistry 52, 147–153. - PubMed
-
- Berger F, Grini PE, Schnittger A. 2006. Endosperm: an integrator of seed growth and development. Current Opinion in Plant Biology 9, 664–670. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

