Development of High Affinity and High Specificity Inhibitors of Matrix Metalloproteinase 14 through Computational Design and Directed Evolution
- PMID: 28087697
- PMCID: PMC5336179
- DOI: 10.1074/jbc.M116.756718
Development of High Affinity and High Specificity Inhibitors of Matrix Metalloproteinase 14 through Computational Design and Directed Evolution
Abstract
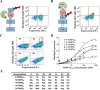
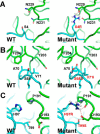
Degradation of the extracellular matrices in the human body is controlled by matrix metalloproteinases (MMPs), a family of more than 20 homologous enzymes. Imbalance in MMP activity can result in many diseases, such as arthritis, cardiovascular diseases, neurological disorders, fibrosis, and cancers. Thus, MMPs present attractive targets for drug design and have been a focus for inhibitor design for as long as 3 decades. Yet, to date, all MMP inhibitors have failed in clinical trials because of their broad activity against numerous MMP family members and the serious side effects of the proposed treatment. In this study, we integrated a computational method and a yeast surface display technique to obtain highly specific inhibitors of MMP-14 by modifying the natural non-specific broad MMP inhibitor protein N-TIMP2 to interact optimally with MMP-14. We identified an N-TIMP2 mutant, with five mutations in its interface, that has an MMP-14 inhibition constant (Ki ) of 0.9 pm, the strongest MMP-14 inhibitor reported so far. Compared with wild-type N-TIMP2, this variant displays ∼900-fold improved affinity toward MMP-14 and up to 16,000-fold greater specificity toward MMP-14 relative to other MMPs. In an in vitro and cell-based model of MMP-dependent breast cancer cellular invasiveness, this N-TIMP2 mutant acted as a functional inhibitor. Thus, our study demonstrates the enormous potential of a combined computational/directed evolution approach to protein engineering. Furthermore, it offers fundamental clues into the molecular basis of MMP regulation by N-TIMP2 and identifies a promising MMP-14 inhibitor as a starting point for the development of protein-based anticancer therapeutics.
Keywords: binding affinity; computational protein design; directed evolution; matrix metalloproteinases; metastasis; protease inhibitor; protein-protein interaction; proteolysis; yeast surface display.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures






References
-
- Brinckerhoff C. E., and Matrisian L. M. (2002) Matrix metalloproteinases: a tail of a frog that became a prince. Nat. Rev. Mol. Cell Biol. 3, 207–214 - PubMed
-
- Vandenbroucke R. E., and Libert C. (2014) Is there new hope for therapeutic matrix metalloproteinase inhibition? Nat. Rev. Drug Discov. 13, 904–927 - PubMed
-
- Seiki M. (1999) Membrane-type matrix metalloproteinases. APMIS 107, 137–143 - PubMed
-
- Dollery C. M., and Libby P. (2006) Atherosclerosis and proteinase activation. Cardiovasc. Res. 69, 625–635 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

