Defining genetic and chemical diversity in wheat grain by 1H-NMR spectroscopy of polar metabolites
- PMID: 28087883
- PMCID: PMC5516129
- DOI: 10.1002/mnfr.201600807
Defining genetic and chemical diversity in wheat grain by 1H-NMR spectroscopy of polar metabolites
Abstract
Scope: The application of high-throughput 1H nuclear magnetic resonance (1H-NMR) of unpurified extracts to determine genetic diversity and the contents of polar components in grain of wheat.
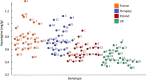
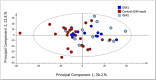
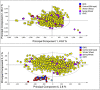
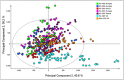
Methods and results: Milled whole wheat grain was extracted with 80:20 D2 O:CD3 OD containing 0.05% d4 -trimethylsilylpropionate. 1H-NMR spectra were acquired under automation at 300°K using an Avance Spectrometer operating at 600.0528 MHz. Regions for individual metabolites were identified by comparison to a library of known standards run under identical conditions. The individual 1H-NMR peaks or levels of known metabolites were then compared by Principal Component Analysis using SIMCA-P software.
Conclusions: High-throughput 1H-NMR is an excellent tool to compare the extent of genetic diversity within and between wheat species, and to quantify specific components (including glycine betaine, choline, and asparagine) in individual genotypes. It can also be used to monitor changes in composition related to environmental factors and to support comparisons of the substantial equivalence of transgenic lines.
Keywords: Grain Composition; Metabolomics; NMR Spectroscopy; Transgenic; Wheat.
© 2017 Rothamsted Research. Molecular Nutrition & Food Research Published by Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures



References
-
- Feldman, M. , in: Smartt J., Simmonds N. W. (Eds.), Evolution of Crop Plants, Longman Scientific and Technical, Harlow, UK: 1995, pp. 185–192.
-
- Mattei, J. , Malik, V. , Wedick, N. M. , Hu, F. B. et al., Reducing the global burden of type 2 diabetes by improving the quality of staple foods: the global nutrition and epidemiologic transition initiative. Global. Health 2015, 11, 23. doi: 10.1186/s12992-015-0109-9. - DOI - PMC - PubMed
-
- Feldman, M. , in: Bonjean A. P., Angus W. J. (Eds.), The World Wheat Book: A History of Wheat Breeding, Lavoisier Publishing, Paris, France: 2001, pp. 3–56.
-
- Khakimov, B. , Bak, S. , Engelsen. S. B. , High‐throughput cereal metabolomics: current analytical technologies, challenges and perspectives. J. Cereal Sci. 2014, 59, 393–418.
Publication types
MeSH terms
Grants and funding
- BB/E015794/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/C/00004975/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/C/00004976/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G004781/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/C/00004158/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

