Molecular characterization of a novel orthomyxovirus from rainbow and steelhead trout (Oncorhynchus mykiss)
- PMID: 28088362
- PMCID: PMC7111338
- DOI: 10.1016/j.virusres.2017.01.005
Molecular characterization of a novel orthomyxovirus from rainbow and steelhead trout (Oncorhynchus mykiss)
Abstract
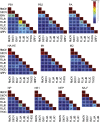
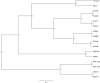
A novel virus, rainbow trout orthomyxovirus (RbtOV), was isolated in 1997 and again in 2000 from commercially-reared rainbow trout (Oncorhynchus mykiss) in Idaho, USA. The virus grew optimally in the CHSE-214 cell line at 15°C producing a diffuse cytopathic effect; however, juvenile rainbow trout exposed to cell culture-grown virus showed no mortality or gross pathology. Electron microscopy of preparations from infected cell cultures revealed the presence of typical orthomyxovirus particles. The complete genome of RbtOV is comprised of eight linear segments of single-stranded, negative-sense RNA having highly conserved 5' and 3'-terminal nucleotide sequences. Another virus isolated in 2014 from steelhead trout (also O. mykiss) in Wisconsin, USA, and designated SttOV was found to have eight genome segments with high amino acid sequence identities (89-99%) to the corresponding genes of RbtOV, suggesting these new viruses are isolates of the same virus species and may be more widespread than currently realized. The new isolates had the same genome segment order and the closest pairwise amino acid sequence identities of 16-42% with Infectious salmon anemia virus (ISAV), the type species and currently only member of the genus Isavirus in the family Orthomyxoviridae. However, pairwise comparisons of the predicted amino acid sequences of the 10 RbtOV and SttOV proteins with orthologs from representatives of the established orthomyxoviral genera and a phylogenetic analysis using the PB1 protein showed that while RbtOV and SttOV clustered most closely with ISAV, they diverged sufficiently to merit consideration as representatives of a novel genus. A set of PCR primers was designed using conserved regions of the PB1 gene to produce amplicons that may be sequenced for identification of similar fish orthomyxoviruses in the future.
Keywords: Fish virus; Oncorhynchus mykiss; Orthomyxovirus; Rainbow trout.
Published by Elsevier B.V.
Figures




References
-
- Bacharach E., Mishra N., Briese T., Zody M.C., Tsofack J.E.K., Zamostiano R., Berkowitz A., Ng J., Nitido A., Corvelo A., Toussaint N.C., Abel Nielsen S.C., Hornig M., Del Pozo J., Bloom T., Ferguson H., Eldar A., Lipkin W.I. Characterization of a novel orthomyxo-like virus causing mass die-offs of tilapia. mBio. 2016;7(2):00431–516. doi: 10.1128/mbio.00431-16. - DOI - PMC - PubMed
-
- Bankevich A., Nurk S., Anstipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. - PMC - PubMed
-
- Batts W.N., Winton J.R. Enhanced detection of infectious hematopoietic necrosis virus and other fish viruses by pretreatment of cell monolayers with polyethylene glycol. J. Aquat. Anim. Health. 1989;1:284–290.
-
- Bouchard D., Keleher W., Opitz H.M., Blake S., Edwards K.C., Nicholson B.L. Isolation of infectious salmon anemia virus (ISAV) from Atlantic salmon in New Brunswick, Canada. Dis. Aquat. Org. 1999;35:131–137. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

