Deciphering the Fringe-Mediated Notch Code: Identification of Activating and Inhibiting Sites Allowing Discrimination between Ligands
- PMID: 28089369
- PMCID: PMC5263050
- DOI: 10.1016/j.devcel.2016.12.013
Deciphering the Fringe-Mediated Notch Code: Identification of Activating and Inhibiting Sites Allowing Discrimination between Ligands
Abstract
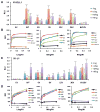
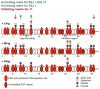
Fringe proteins are β3-N-acetylglucosaminyltransferases that modulate Notch activity by modifying O-fucose residues on epidermal growth factor-like (EGF) repeats of Notch. Mammals have three Fringes: Lunatic, Manic, and Radical. While Lunatic and Manic Fringe inhibit Notch1 activation from Jagged1 and enhance activation from Delta-like 1, Radical Fringe enhances signaling from both. We used a mass spectrometry approach to determine whether the variable effects of Fringes on Notch1 result from generation of unique glycosylation patterns on Notch1. We found that Lunatic and Manic Fringe modified similar sites on Notch1, while Radical Fringe modified a subset. Fringe modifications at EGF8 and EGF12 enhanced Notch1 binding to and activation from Delta-like 1, while modifications at EGF6 and EGF36 (added by Manic and Lunatic but not Radical) inhibited Notch1 activation from Jagged1. Combined, these results suggest that Fringe modifications "mark" different regions in the Notch1 extracellular domain for activation or inhibition.
Keywords: EGF repeats; Fringe; Notch; O-fucose; development; glycosylation; signal transduction.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


References
-
- Benedito R, Roca C, Sorensen I, Adams S, Gossler A, Fruttiger M, Adams RH. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135. - PubMed
-
- Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–415. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

