Coupling of Homologous Recombination and the Checkpoint by ATR
- PMID: 28089683
- PMCID: PMC5496772
- DOI: 10.1016/j.molcel.2016.12.007
Coupling of Homologous Recombination and the Checkpoint by ATR
Abstract
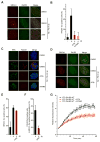
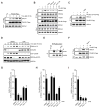
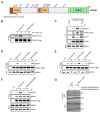
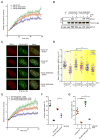
ATR is a key regulator of cell-cycle checkpoints and homologous recombination (HR). Paradoxically, ATR inhibits CDKs during checkpoint responses, but CDK activity is required for efficient HR. Here, we show that ATR promotes HR after CDK-driven DNA end resection. ATR stimulates the BRCA1-PALB2 interaction after DNA damage and promotes PALB2 localization to DNA damage sites. ATR enhances BRCA1-PALB2 binding at least in part by inhibiting CDKs. The optimal interaction of BRCA1 and PALB2 requires phosphorylation of PALB2 at S59, an ATR site, and hypo-phosphorylation of S64, a CDK site. The PALB2-S59A/S64E mutant is defective for localization to DNA damage sites and HR, whereas the PALB2-S59E/S64A mutant partially bypasses ATR for its localization. Thus, HR is a biphasic process requiring both high-CDK and low-CDK periods. As exemplified by the regulation of PALB2 by ATR, ATR promotes HR by orchestrating a "CDK-to-ATR switch" post-resection, directly coupling the checkpoint to HR.
Keywords: ATR; BRCA1; CDK; PALB2; checkpoint; homologous recombination.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
References
-
- Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

